Volume 24, Number 12—December 2018
Research
Crimean-Congo Hemorrhagic Fever Virus, Mongolia, 2013–2014
Figure 2
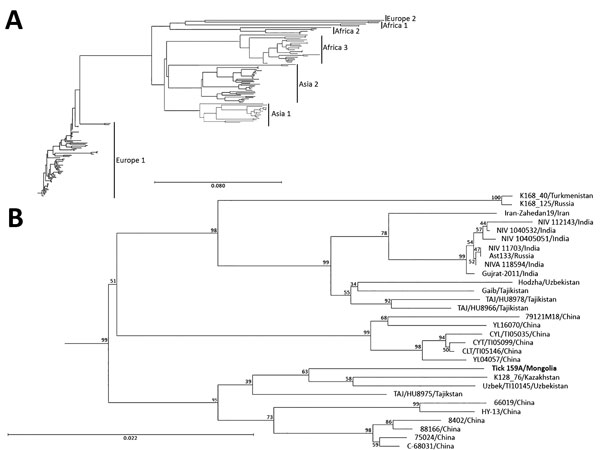
Figure 2. Phylogenetic characterization of partial small (S) segment sequence of Crimean-Congo hemorrhagic fever virus (CCHFV) isolate from tick pool 159A, Mongolia, 2013–2014. Near full–length CCHFV S segments from GenBank were aligned with the S segment sequence from tick pool 159A and a phylogenetic tree was generated. A) Genetic clusters are displayed as previously described (23). B) Detailed view of phylogenetic tree of Asia 2 lineage. S segment of the CCHFV isolate from this study (tick 159A/Mongolia; bold) clusters in the Asia 2 lineage. Scale bars indicate nucleotide substitutions per site.
References
- Bente DA, Forrester NL, Watts DM, McAuley AJ, Whitehouse CA, Bray M. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res. 2013;100:159–89. DOIPubMedGoogle Scholar
- Lukashev AN, Klimentov AS, Smirnova SE, Dzagurova TK, Drexler JF, Gmyl AP. Phylogeography of Crimean Congo hemorrhagic fever virus. PLoS One. 2016;11:e0166744. DOIPubMedGoogle Scholar
- Messina JP, Pigott DM, Golding N, Duda KA, Brownstein JS, Weiss DJ, et al. The global distribution of Crimean-Congo hemorrhagic fever. Trans R Soc Trop Med Hyg. 2015;109:503–13. DOIPubMedGoogle Scholar
- Hewson R, Gmyl A, Gmyl L, Smirnova SE, Karganova G, Jamil B, et al. Evidence of segment reassortment in Crimean-Congo haemorrhagic fever virus. J Gen Virol. 2004;85:3059–70. DOIPubMedGoogle Scholar
- Mostafavi E, Chinikar S, Moradi M, Bayat N, Meshkat M, Fard MK, et al. A case report of crimean congo hemorrhagic Fever in ostriches in iran. Open Virol J. 2013;7:81–3. DOIPubMedGoogle Scholar
- Lindeborg M, Barboutis C, Ehrenborg C, Fransson T, Jaenson TG, Lindgren PE, et al. Migratory birds, ticks, and crimean-congo hemorrhagic fever virus. Emerg Infect Dis. 2012;18:2095–7. DOIPubMedGoogle Scholar
- Shepherd AJ, Swanepoel R, Leman PA, Shepherd SP. Field and laboratory investigation of Crimean-Congo haemorrhagic fever virus (Nairovirus, family Bunyaviridae) infection in birds. Trans R Soc Trop Med Hyg. 1987;81:1004–7. DOIPubMedGoogle Scholar
- Mild M, Simon M, Albert J, Mirazimi A. Towards an understanding of the migration of Crimean-Congo hemorrhagic fever virus. J Gen Virol. 2010;91:199–207. DOIPubMedGoogle Scholar
- Papa A, Tsergouli K, Tsioka K, Mirazimi A. Crimean-Congo hemorrhagic fever: tick-host-virus interactions. Front Cell Infect Microbiol. 2017;7:213. DOIPubMedGoogle Scholar
- Ergönül O. Crimean-Congo haemorrhagic fever. Lancet Infect Dis. 2006;6:203–14. DOIPubMedGoogle Scholar
- Swanepoel R, Gill DE, Shepherd AJ, Leman PA, Mynhardt JH, Harvey S. The clinical pathology of Crimean-Congo hemorrhagic fever. Rev Infect Dis. 1989;11(Suppl 4):S794–800. DOIPubMedGoogle Scholar
- Chumikhin S, Potshild E, Nyamadava P, Peshetnikov I, Tkachenko E, Dzagurova T. Serological and virological tests used for the detection of arboviruses in wild mammals and birds of Mongolia. Med Parazitol (Mosk). 1987:10–2.
- Morikawa S. Serological surveillance of Crimean-Congo haemorrhagic fever infection among sheep in Mongolia. Tokyo: National Institute of Infectious Diseases; 2013.
- Guo R, Shen S, Zhang Y, Shi J, Su Z, Liu D, et al. A new strain of Crimean-Congo hemorrhagic fever virus isolated from Xinjiang, China. Virol Sin. 2017;32:80–8. DOIPubMedGoogle Scholar
- Sun S, Dai X, Aishan M, Wang X, Meng W, Feng C, et al. Epidemiology and phylogenetic analysis of crimean-congo hemorrhagic fever viruses in xinjiang, china. J Clin Microbiol. 2009;47:2536–43. DOIPubMedGoogle Scholar
- Yen YC, Kong LX, Lee L, Zhang YQ, Li F, Cai BJ, et al. Characteristics of Crimean-Congo hemorrhagic fever virus (Xinjiang strain) in China. Am J Trop Med Hyg. 1985;34:1179–82. DOIPubMedGoogle Scholar
- Papa A, Weber F, Hewson R, Weidmann M, Koksal I, Korukluoglu G, et al. Meeting report: First International Conference on Crimean-Congo hemorrhagic fever. Antiviral Res. 2015;120:57–65. DOIPubMedGoogle Scholar
- Garrison AR, Alakbarova S, Kulesh DA, Shezmukhamedova D, Khodjaev S, Endy TP, et al. Development of a TaqMan minor groove binding protein assay for the detection and quantification of Crimean-Congo hemorrhagic fever virus. Am J Trop Med Hyg. 2007;77:514–20. DOIPubMedGoogle Scholar
- Koehler JW, Delp KL, Hall AT, Olschner SP, Kearney BJ, Garrison AR, et al. Sequence optimized real-time reverse transcription polymerase chain reaction assay for detection of Crimean-Congo hemorrhagic fever virus. Am J Trop Med Hyg. 2018;98:211–5. DOIPubMedGoogle Scholar
- Morikawa S. Laboratory diagnosis of Crimean-Congo hemorrhagic fever (CCHF). Tokyo: National Institute of Infectious Diseases; 2012. p. 1–15.
- Deyde VM, Khristova ML, Rollin PE, Ksiazek TG, Nichol ST. Crimean-Congo hemorrhagic fever virus genomics and global diversity. J Virol. 2006;80:8834–42. DOIPubMedGoogle Scholar
- Koehler JW, Delp KL, Kearney BJ, Conrad TA, Schoepp RJ, Garrison AR, et al. Draft genome sequences of eight Crimean-Congo hemorrhagic fever virus strains. Genome Announc. 2017;5:e00240-17. DOIPubMedGoogle Scholar
- Anagnostou V, Papa A. Evolution of Crimean-Congo hemorrhagic fever virus. Infect Genet Evol. 2009;9:948–54. DOIPubMedGoogle Scholar
- Spengler JR, Bergeron É, Rollin PE. Seroepidemiological studies of Crimean-Congo hemorrhagic fever virus in domestic and wild animals. PLoS Negl Trop Dis. 2016;10:e0004210. DOIPubMedGoogle Scholar
- Xia H, Li P, Yang J, Pan L, Zhao J, Wang Z, et al. Epidemiological survey of Crimean-Congo hemorrhagic fever virus in Yunnan, China, 2008. Int J Infect Dis. 2011;15:e459–63. DOIPubMedGoogle Scholar
Page created: November 20, 2018
Page updated: November 20, 2018
Page reviewed: November 20, 2018
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.