Rickettsial Illnesses as Important Causes of Febrile Illness in Chittagong, Bangladesh
Hugh W. Kingston, Mosharraf Hossain, Stije Leopold, Tippawan Anantatat, Ampai Tanganuchitcharnchai, Ipsita Sinha, Katherine Plewes, Richard J. Maude, M.A. Hassan Chowdhury, Sujat Paul, Rabiul Alam Mohammed Erfan Uddin, Mohammed Abu Naser Siddiqui, Abu Shahed Zahed, Abdullah Abu Sayeed, Mohammed Habibur Rahman, Anupam Barua, Mohammed Jasim Uddin, Mohammed Abdus Sattar, Arjen M. Dondorp, Stuart D. Blacksell, Nicholas P.J. Day, Aniruddha Ghose, Amir Hossain, and Daniel H. Paris

Author affiliations: Charles Darwin University, Casuarina, Northern Territory, Australia (H.W. Kingston); Mahidol University, Bangkok, Thailand (H.W. Kingston, S. Leopold, T. Anantatat, A. Tanganuchitcharnchai, I. Sinha, K. Plewes, R.J. Maude, A.M. Dondorp, S.D. Blacksell, N.P.J. Day, D.H. Paris); Chittagong Medical College Hospital, Chittagong, Bangladesh (M. Hossain, M.A.H. Chowdhury, S. Paul, R.A.M.E. Uddin, M.A.N. Siddiqui, A.S. Zahed, A.A. Sayeed, M.H. Rahman, A. Barua, M.J. Uddin, M.A. Sattar, A. Ghose, A. Hossain); Oxford University, Oxfordshire, UK (I. Sinha, R.J. Maude, A.M. Dondorp, S.D. Blacksell, N.P.J. Day, D.H. Paris); Harvard University, Boston, Massachusetts, USA (R.J. Maude); Swiss Tropical and Public Health Institute, Basel, Switzerland (D.H. Paris); University of Basel, Basel (D.H. Paris)
Main Article
Figure 3
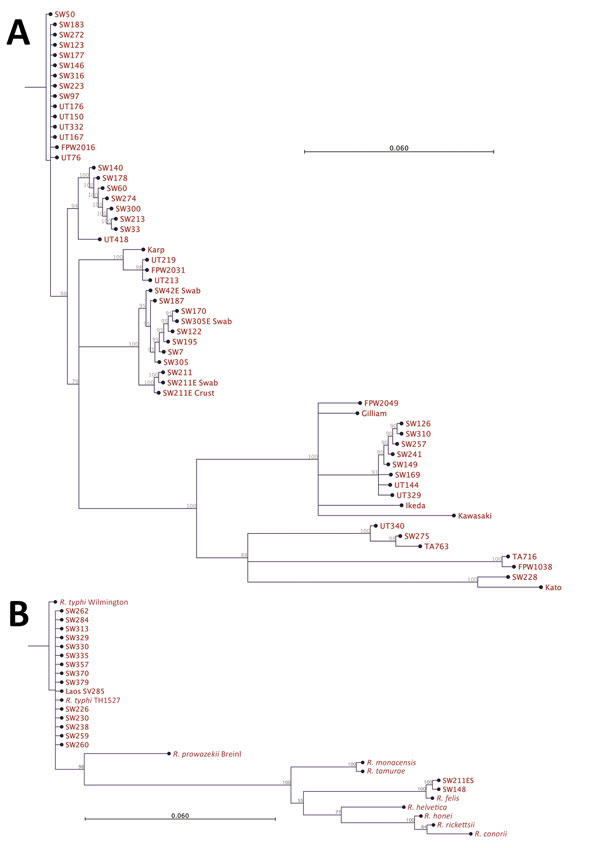
Figure 3. Phylogenetic analysis of pathogens contributing to rickettsial illnesses, Chittagong Medical College Hospital, Chittagong, Bangladesh, August 2014–September 2015. A) Phylogenetic dendrogram based on the nucleotide sequence of the partial open reading frame of the 56-kDa TSA gene (aligned and cropped to ≈450 bp), depicting Orientia tsutsugamushi strains in relationship with reference and other strains. O. tsutsugamushi genotypes in Bangladesh included Karp, Gilliam, Kato, and TA763 strains, with a predominance of Karp-like strains. B) Rickettsia spp. as characterized by 17-kDa gene sequencing (aligned and cropped to 314 bp). The predominant pathogen identified was R. typhi. Of note, 2 R. felis infection cases were identified, including 1 systemic bloodstream infection and 1 scrub typhus case with eschar swab positivity in patient no. SW211ES (Technical Appendix Table), whose blood specimen was negative for R. felis, suggesting possible skin colonization of R. felis. Scale bar indicates nucleotide substitutions per site; branches shorter than 0.002 are shown as having a length of 0.002.
Main Article
Page created: March 16, 2018
Page updated: March 16, 2018
Page reviewed: March 16, 2018
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
