Volume 24, Number 4—April 2018
Dispatch
Direct Whole-Genome Sequencing of Cutaneous Strains of Haemophilus ducreyi
Figure 2
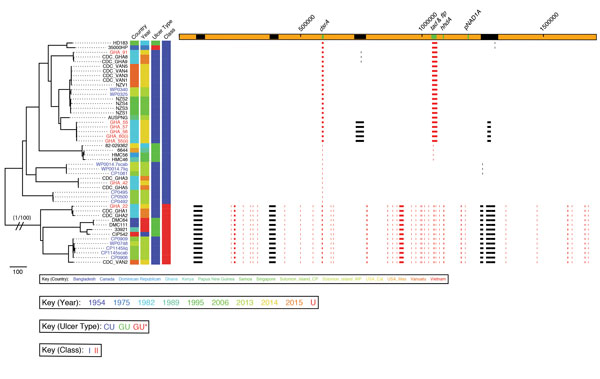
Figure 2. Phylogenetic tree of Haemophilus ducreyi genome sequences inferred from mapping using the H. ducreyi 35000HP strain as reference and after removing high-density single-nucleotide polymorphisms regions with Gubbins (3). Included are published genomes (black text), Ghanaian strains (gray text, GHA designations), and Solomon Islands strains (gray text, CP/WP designations). Sequences from cutaneous ulcers in Ghana and the Solomon Islands were found within both previously described clades of H. ducreyi class I and class II. Scale bar indicates nucleotide substitutions per site. An expanded version of this figure providing complete phylogeny details, including countries of origin, years, ulcer types, and genome region designations, is provided in Technical Appendix 1 Figure.
1These authors contributed equally to this article.