Volume 24, Number 9—September 2018
Dispatch
Aortic Endograft Infection with Mycobacterium chimaera and Granulicatella adiacens, Switzerland, 2014
Figure 2
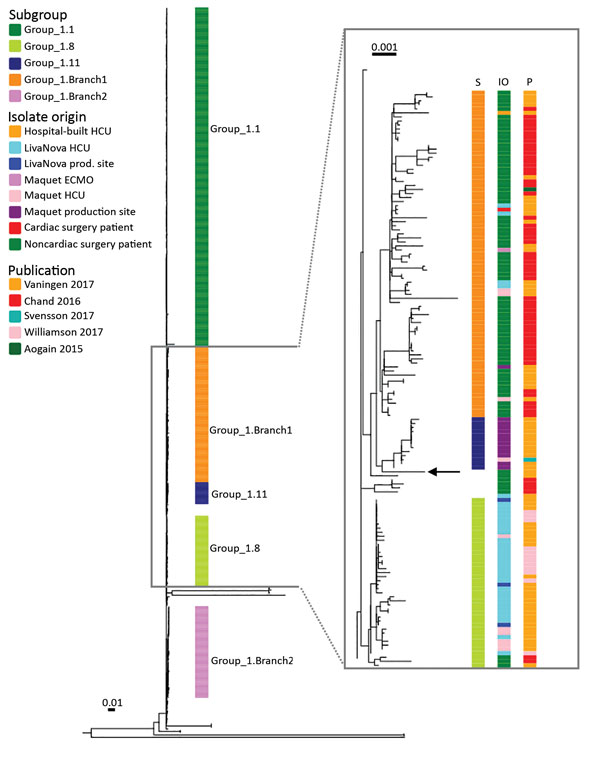
Figure 2. Phylogeny of isolate from case-patient who received an abdominal aortic endograft and was later diagnosed with Mycobacterium chimaera and Granulicatella adiacens infection, Switzerland, 2014, and comparison isolates. Maximum-likelihood tree was built from 14,192 single-nucleotide polymorphism positions of 437 group 1 Mycobacterium chimaera isolates mapped to the DSM-44623 M. chimaera genome (GenBank accession no. NZ_CP015278.1). DSM-44623 is shown as a rectangular phylogram with the inferred subgroups indicated. Inset box shows subgroups 1.8, 1.11, and 1.Branch1, annotated with isolate origin and the source publication. Black arrow indicates position of the patient isolate. Group 1.11 consisted mainly of samples collected at the Maquet production site in Rastatt, Germany (n = 12); 1 isolate came from an in-use Maquet HCU. Branch 1 contained primarily strains from patients with pulmonary M. chimaera infections (n = 70) and strains from LivaNova HCUs (n = 4), Maquet HCUs (n = 3), Maquet ECMOs (n = 11), a hospital-built HCU (n = 1), Maquet production site (n = 1), and a patient infected after cardiac surgery (n = 1). ECMO, extracorporeal membrane oxygenation; HCU, heater–cooler unit. Scale bars indicate numbers of substitutions per site.