Volume 24, Number 9—September 2018
Research Letter
Rubella Virus Genotype 1E in Travelers Returning to Japan from Indonesia, 2017
Figure
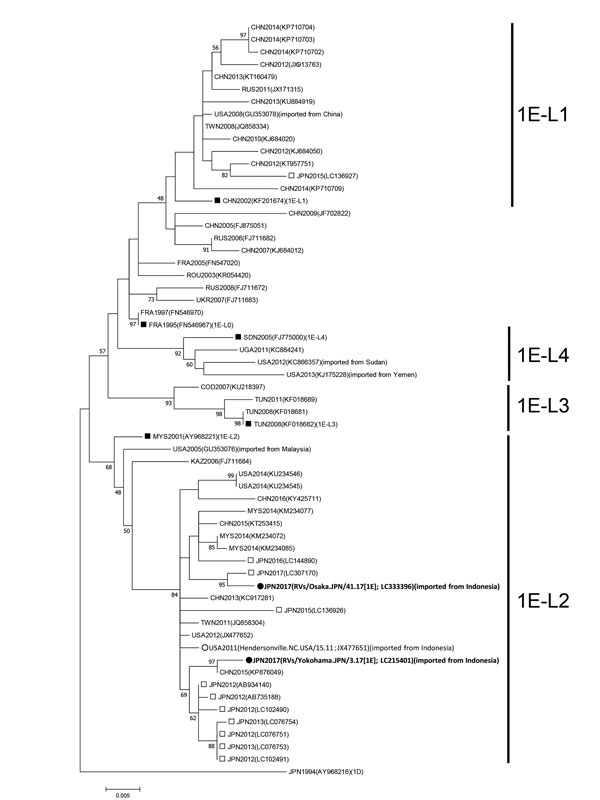
Figure. Maximum-likelihood phylogram of the molecular window region (739 nt) within the E1 gene of rubella virus genotype 1E from a 29-year-old man in Japan who had traveled to Indonesia (black circles). We constructed a phylogenetic tree using 61 strains, including the genotype reference strains and the candidate lineage reference strains, using MEGA version 7.0 (http://www.megasoftware.net) and the Tamura-Nei model. Numbers at nodes indicate the bootstrap support values, given as a percentage of 1,000 replicates (values <45 are omitted). The genotype 1D reference strain (RVi/Saitama.JPN/0.94/[1D]) is included as an outgroup. White circles indicate the genotype 1E strain detected in patients returning to the United States from Indonesia in 2011. Black squares indicate candidate genotype 1E lineage reference strains and genotype 1E strains. White squares indicate the strains detected in Japan from 2012–2017. Each strain identification consists of a 3-letter country name abbreviation and detection year. Accession numbers are shown in parentheses. Scale bar indicates nucleotide substitutions per site. COD, Democratic Republic of the Congo; FRA, France; JPN, Japan; KAZ, Kazakhstan; CHN, China; MYS, Malaysia; ROU, Romania; RUS, Russia; SDN, Sudan; TWN, Taiwan; TUN, Tunisia; UGA, Uganda; UKR, Ukraine; USA, United States of America.