Volume 25, Number 1—January 2019
Research
Risk Factors for Elizabethkingia Acquisition and Clinical Characteristics of Patients, South Korea
Cite This Article
Citation for Media
Abstract
Elizabethkingia infections are difficult to treat because of intrinsic antimicrobial resistance, and their incidence has recently increased. We conducted a propensity score–matched case–control study during January 2016–June 2017 in South Korea and retrospectively studied data from patients who were culture positive for Elizabethkingia species during January 2009–June 2017. Furthermore, we conducted epidemiologic studies of the hospital environment and mosquitoes. The incidence of Elizabethkingia increased significantly, by 432.1%, for 2016–2017 over incidence for 2009–2015. Mechanical ventilation was associated with the acquisition of Elizabethkingia species. Because Elizabethkingia infection has a high case-fatality rate and is difficult to eliminate, intensive prevention of contamination is needed.
The genus Elizabethkingia comprises glucose-nonfermenting, gram-negative rods that are widely distributed in natural environments, including in soil and freshwater, and in hospital environments (1). E. meningoseptica (originally named Chryseobacterium meningoseptica) has been associated with opportunistic infections, such as sepsis in immunocompromised patients and meningitis in neonates (2). Two new species of Elizabethkingia have been proposed: E. miricola, which was first isolated from water from the Russian space station MIR in 2003 (3,4); and E. anophelis, which was first isolated from the midgut of the Anopheles gambiae mosquito in 2011 (5). Because E. anophelis was the most frequently isolated Elizabethkingia species in recent clinical studies, as confirmed by 16s rRNA gene sequencing (6,7), but is commonly misidentified as E. meningoseptica, many previously reported cases of E. meningoseptica could actually have been caused by E. anophelis (8,9).
Infection caused by Elizabethkingia species is difficult to treat and results in a high case-fatality rate, probably because of intrinsic antimicrobial resistance (10). E. meningoseptica has been documented to carry class A extended-spectrum β-lactamases and 2 chromosomal metallo-β-lactamases (11,12).
Some outbreaks of Elizabethkingia species have been reported to have resulted from a contaminated water source (13–15). Furthermore, recent increases in the annual incidence of Elizabethkingia species. have been reported in many countries (14,16–19). However, knowledge about host risk factors associated with the acquisition of Elizabethkingia species is lacking, and no evidence exists that mosquitoes or other sources act as vectors in transmitting it to humans. Thus, we investigated the annual incidence and clinical characteristics of Elizabethkingia acquisition in a tertiary teaching hospital in Seoul, South Korea. We aimed to determine whether the incidence of Elizabethkingia species had increased in this hospital and to analyze the risk factors associated with Elizabethkingia acquisition. To identify the source of Elizabethkingia, we obtained and analyzed epidemiologic studies from the hospital environment and mosquitoes.
Study Participants
We retrospectively collected data from all nonduplicate persons who had positive culture results for Elizabethkingia species from Severance Hospital, a >2,000-bed tertiary teaching hospital in South Korea, during January 1, 2009–June 30, 2017. The hospital had 10 intensive care units (ICUs) for adults and 2 for children. During this period, the annual number of inpatient-days was >670,000.
Because we had identified strains in our previous study (7), we updated our in-house library of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MS) (Bruker Daltonic GmbH, https://www.bruker.com). Elizabethkingia species were identified by 2 matrix-assisted laser desorption/ionization time-of-flight MS systems; the Bruker MS used the updated in-house library and the Vitek MS (bioMèrieux, https://www.biomerieux.com) used the latest version of IVD (in vitro diagnostic database) V3.2. Strains with discrepant results were confirmed by 16S rRNA gene sequencing using universal primers.
We collected the following clinical data using electronic medical records: age-adjusted Charlson comorbidity index (20), sex, sites of specimen collection, date of specimen collection, date of patient death, pulse rate, oxygen saturation, body temperature, chest radiograph results, and any antimicrobial agents administered during hospitalization. We also obtained available laboratory findings from the same day as specimen collection and within 7 days from the same day as specimen collection, including C-reactive protein level, erythrocyte sedimentation rate, and leukocyte count. The Institutional Review Board at Severance Hospital, affiliated with Yonsei University Health System (2017–2101–001), approved this study.
Epidemiologic Study of Environmental Sources and Mosquitoes
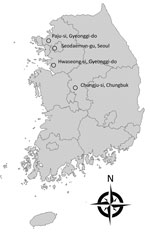
We obtained extensive surveillance cultures of 281 common environmental sources by swab culture of equipment and surfaces within patient rooms, restrooms, nursing stations, electronics, furniture, patient care devices, patient transport carts, sinks, and water taps. Additionally, during July–September 2017, we collected adult mosquitoes in 1 urban site (Seodaemun-gu, Seoul, where Severance Hospital is located) and 3 rural areas (Hwaseong-si, Gyeonggi-do; Paju-si, Gyeonggi-do; and Chungju-si, Chungbuk, where annual zoonotic disease monitoring had taken place for their dense population of animal farms) (Figure 1). Mosquitoes were collected using Insect Light Traps Model SR-2000 (Sin Young Inc., Seoul, South Korea) and identified under a stereomicroscope after cold anesthesia, as in the previous study (21).
All swab samples and midguts of mosquitoes were inoculated on sheep blood agar and MacConkey agar and incubated overnight. DNA was extracted from mosquitoes using a simple boiling method, and PCR was performed using Elizabethkingia species–specific primers (forward, GAACACGTGTGCAACCTGCC; reverse, TCCAGCCACTTCAACCTTAC) and the following cycle parameters: 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 58°C for 2 min, and 72°C for 1 min; followed by a final extension step at 72°C for 7 min (22).
Pulsed-Field Gel Electrophoresis
We conducted pulsed-field gel electrophoresis (PFGE) analysis of XbaI-digested isolated chromosomal DNA from a total of 12 strains isolated from the environment (7 E. anophelis, 3 E. miricola, 2 E. meningoseptica) and 54 stored strains isolated from inpatients since 2017. PFGE patterns were analyzed using the CHEF DR II system (Bio-Rad, http://www.bio-rad.com) as previously described (9).
Definition
We defined true pathogen cases according to the definitions of Moore et al. and the Centers for Disease Control and Prevention. In brief, we defined these cases as patients with the monomicrobial acquisition of Elizabethkingia species and 1 of the following parameters within 2 days before and after acquisition without any other recognized cause: body temperature <36°C or >38°C, pulse rate >90 beats/min (reference range 60–100 beats/min), leukocyte count <4 or >12 × 109 cells/L (reference range 4.0–10.9 × 109 cells/L), C-reactive protein >100 mg/L (reference range 0–8 mg/L), or chest radiography showing new pulmonary infiltrations (13,23). According to previous studies, outbreaks are determined by whether they exceed 2 SD of the previous disease incidence (24,25).
Propensity Score–Matched Analysis
After 2016, the incidence of isolated Elizabethkingia species increased significantly. To analyze the increased incidence, we conducted surveillance culture study and compared clinical characteristics of patients who acquired Elizabethkingia species before and after 2016. In both the surveillance culture study and the statistical analysis, only 3 study wards showed positive results for the acquisition of Elizabethkingia: an ICU (ICU 1, 18 beds) used for cardiovascular disease, an isolation ward (ward A, 50 beds) used for patients with vancomycin-resistant Enterococcus, and a general ward (ward B, 50 beds) used for pulmonary disease patients. A total of 6,583 patients have been hospitalized in these wards since 2016.
To adjust confounding factors for the acquisition of Elizabethkingia species, we conducted a propensity score (PS)–matched case–control study. We defined case-patients as patients with Elizabethkingia species isolated from any clinical specimens during January 2016–June 2017 in ICU 1, ward A, or ward B and control patients as patients without Elizabethkingia species in these 3 study wards. We selected 3 variables—hospital ward (p<0.001), period of admission (p = 0.041), and length of stay in the 3 study wards (p<0.001)—for adjustment by univariate analysis (26,27). We estimated a PS for the predicted probability of acquisition of Elizabethkingia species in each patient using the logistic regression model. Then we performed a PS-matched analysis by attempting to match case-patients and control patients (1:3 match) using the nearest-neighbor-matching method. A match occurred when the difference in the logits of the PS was <0.2 times the SD of the scores.
Statistical Analysis
We assessed all variables using the Shapiro-Wilk test to evaluate Gaussian distributions. Descriptive statistics are presented as a median and interquartile range (IQR) for continuous variables or numbers and percentage for categorical variables. Comparisons between groups were analyzed using the Mann-Whitney U test for continuous variables and the Fisher exact test for categorical variables.
Using conditional logistic regression, we conducted univariate and multivariate regressions between case-patients and control patients of the 3 study wards. Dependent variables included in the multivariate analysis were selected based on statistical significance provided by univariate analysis. Incidence rate ratios and 95% CIs were calculated by comparing the mean incidences between 2009–2015 and 2016–2017 by Poisson regression.
All reported p values are 2-tailed, and p values <0.05 indicate statistical significance. We conducted statistical analyses using R statistical software (R Studio, Inc., https://www.r-project.org).
The annual incidence of Elizabethkingia acquisition in Severance Hospital increased in 2011 (Table 1; Figure 2). According to the defined threshold, years with incidence >2 SD were 2011, 2012, 2013, 2014, and 2016. In particular, incidence increased most significantly to 109.82 cases/1 million inpatient-days in 2016 (p<0.001). An additional 50 cases were reported during January–June 2017, which corresponded to 127.79 cases/1 million inpatient-days. The acquisition incidence of Elizabethkingia species increased significantly, by 432.1%, during 2016–2017 over the acquisition incidence during 2009–2015 (relative risk [RR] 4.17, 95% CI 3.28–5.29; p<0.001), mainly because of the increase in strains isolated from respiratory specimens (incidence rate ratio 3.22, 95% CI 2.46–4.20; p<0.001).
We identified 269 patients who acquired Elizabethkingia species during January 2009–June 2017, of whom 134 (49.8%) were reported during 2016–June 2017 (Table 2). Patients who acquired Elizabethkingia during 2016–June 2017 were more frequently classified as having contracted a nosocomial infection than were patients who acquired Elizabethkingia during 2009–2015. The number of cases in the 3 study wards increased significantly but not in the other wards. In addition, more patients with chronic pulmonary disease or diabetes mellitus were seen during 2016–2017. More patients had a history of mechanical ventilation, a longer length of hospital stay, and a history of steroid use during 2009–2015 than during 2016–2017.
Surveillance Studies
We isolated 12 Elizabethkingia strains; all were derived from the 3 study wards. Seven E. anophelis isolates (4 from water taps in ICU 1, 2 from washbasins in ICU 1, and 1 from the suction regulator in ward A), 3 E. miricola isolates (3 from washbasins in ward A), and 2 E. meningoseptica isolates (1 from the mechanical ventilator after removal from a patient and 1 from the suction regulator in ward B) were identified by surveillance cultures.
We conducted PFGE typing on 54 clinical isolates (40 E. anophelis, 10 E. miricola, and 4 E. meningoseptica) from inpatients and 7 E. anophelis, 3 E. miricola, and 2 E. meningoseptica isolates from environmental samples. PFGE patterns showed that E. anophelis isolates belonged to 8 different PFGE groups, E. miricola to 4 groups, and E. meningoseptica to 2 groups (Figure 3). Five patients with E. anophelis (1 patient from ward B and 4 patients from other locations) had a history of admission to ICU 1 (Figure 3, panel A). Of patients from other locations, 1 had a history of admission to ward A and 2 had histories of admission to ward B. One patient in ICU 1 had moved from ward A, where the major cluster of environmental samples was isolated (Figure 3, panel B). Similarly, 1 patient in ICU 1 was transferred from ward B (Figure 3, panel C). This patient’s history of ward transfers suggests that transmission of the bacteria from patient to patient might be a cause of spreading. However, we cannot rule out the existence of other environmental sources.
We conducted PCR on 30 Anopheles sinensis, 8 Culex tritaeniorhynchus, and 3 Aedes vexans mosquitoes and on surveillance cultures collected from the midgut of mosquitoes. All yielded negative results.
Epidemiologic Results Before and After PS Matching
Of the 6,583 patients potentially exposed on the 3 study wards, 74 were colonized or infected with Elizabethkingia species (Table 3). Case-patients and control-patients differed significantly in the proportion of hospitalized wards. The case-patient group, had higher admission rates to ward A and ward B, whereas control-patients had a higher rate of admission to ICU 1. Furthermore, case-patients had a significantly longer stay in the 3 study wards than did control-patients. In the 3 study wards, case-patients spent a median of 55 days (IQR 20–131 days), significantly longer than that of control patients (3 days [IQR 2–8 days]; p<0.001).
We conducted PS matching to adjust baseline demographics and clinical variables between the case-patient and control-patient groups. PS matching resulted in 52 matched pairs at a 1:3 ratio. After matching, we included 52 of 74 case-patients in the analysis. Confounding variables were well balanced in the 2 groups, including hospitalization ward and the period of admission and length of stay in the 3 study wards. Furthermore, the 2 comorbidities after PS matching did not differ significantly because there was an adjustment to the proportions of admission to ward A, which had high rates of patients with hematologic malignancy, and ward B, which had high rates of patients with chronic pulmonary disease. However, use of mechanical ventilation still differed significantly after PS matching (p<0.001).
We used univariate and multivariate analyses with conditional logistic regression to identify independent risk factors (Table 4). Only use of mechanical ventilation (adjusted odds ratio [OR] 50.44 [95% CI 6.74–377.48]; p<0.001) was associated with the acquisition of Elizabethkingia species.
Only 30 patients were classified as true pathogen cases (Table 5). In the hospital mortality group, the median total hospitalization stay was longer than that of the nonhospital mortality group (77.5 vs. 38.5 days; p = 0.04). More case-patients were treated with carbapenem or trimethoprim/sulfamethoxazole than were those in the nonhospital mortality group (5 vs. 2 case-patients; p = 0.05).
The incidence of infection with Elizabethkingia species has increased in recent years in many countries (14,16–19). Furthermore, a large-scale outbreak was reported in community settings in the United States (28).
In previous studies, the reported annual incidence of E. meningoseptica ranged from 0.007 to 0.399 cases per 1,000 admissions (19,29). We reported the antimicrobial resistance mechanisms and susceptibility rates of Elizabethkingia species isolated from Severance Hospital in 2010 (11) and 2016 (7). Recently, the incidence of isolation in this hospital increased significantly, from 0.02 to 0.88 per 1,000 admissions during 2009–2017, mainly in the 3 study wards.
We analyzed the risk factors associated with the acquisition of Elizabethkingia species after controlling for other confounding variables. Using multivariate analysis, we found that the probability of acquiring Elizabethkingia species was significantly influenced by whether a patient received mechanical ventilation, even after PS matching and adjustment for other variables. Although some previous studies have suggested that mechanical ventilators could be a risk factor for colonization or infection with Elizabethkingia species, they did not provide a statistical analysis and included only a small number of patients (16,30). In our study, a total of 214 (79.6%) patients from among the 269 patients seen during January 2009–June 2017 received mechanical ventilation, as did 72 (97.3%) of 74 case-patients admitted to the 3 study wards during January 2016–June 2017. We included a large number of cases and tried to control for confounding variables using PS matching, thus ensuring that mechanical ventilation is related to the acquisition of Elizabethkingia species.
Water or water-related equipment can serve as a waterborne pathogen reservoir in the hospital environment (31). Previous studies also have associated a water source with acquisition of Elizabethkingia because of the bacterium’s ability to form a biofilm in moist environments. Balm et al. reported the infections of 5 patients in 1 outbreak with E. meningoseptica were related to a hand hygiene sink aerator (14). Moore et al. identified 30 patients as having acquired E. meningoseptica during an outbreak; at least 10 of these infections were associated with 5 environmental samples isolated from sinks (13). Elizabethkingia can spread from a humid environment to the surface of medical devices or dry materials by the hands of hospital staff or patients (32). In our current study, all 12 environmental isolates shared identical PFGE patterns with clinical isolates. Our finding is consistent with prior reports that Elizabethkingia acquisition might be related to water sources within the hospital environment. In contrast, we could not find any evidence that the local mosquitoes of South Korea act as vehicles of Elizabethkingia transmission.
As extended-spectrum β-lactamase–producing bacteria have increased, the use of carbapenems has inevitably increased. Previous reports have suggested that antimicrobial selective pressure may increase the prevalence of bacteria with natural resistance to carbapenems, such as Elizabethkingia species (33,34). Unlike in our univariate analysis, our multivariate analysis showed no association between antimicrobial exposure to carbapenem and the acquisition of Elizabethkingia. One possible explanation for this finding could be that host factors are a more important selection factor than the antimicrobial drug in the selection of this strain. Another explanation could be that our data lacked the statistical power to detect differences in Elizabethkingia acquisition by antimicrobial exposure to carbapenem.
Difficulties in eradicating and terminating outbreaks of Elizabethkingia caused by a strong biofilm biotype have been reported (13,14,35). Furthermore, the failure of 1,000-ppm sodium hypochlorite and posthandwashing alcohol gel has been documented (34). The acquisition risk can be reduced by regular sink flushing and improvements to the workflow that minimize contamination (13). Fortunately, we succeeded in eliminating Elizabethkingia species in ward B in September 2017. After all structures from which bacteria were isolated were dismantled, the outer surface and inner spaces were cleaned. A sheer force was applied using a sodium hypochlorite solution and a brush, then structures were reinstalled. In ward B, no new patient acquisitions of Elizabethkingia occurred after this effort. These findings also support the possibility that Elizabethkingia acquisition may be related to water source and the contaminated devices. However, a previous study documented that Elizabethkingia species have been re-isolated after a month, even after all contaminated devices were replaced (14). Continuous monitoring, including surveillance culture systems and education for medical staff, may be more important than decontamination in reducing the acquisition of and infection with Elizabethkingia.
Among our data on treatment outcomes for true pathogen cases, none of the antimicrobial agents used after reporting the culture results were related to reducing in-hospital death. Alternatively, patients treated with carbapenem or trimethoprim/sulfamethoxazole had significantly higher in-hospital death rates. However, this analysis included only 30 true pathogen cases, and results could not be adjusted for other potential confounding factors.
The retrospective and single-center nature of the study limited our results. Thus, selection bias might exist in the tests performed on environmental samples and mosquito samples, and we could not identify the species of bacteria that were not stored. It is also difficult to perform additional surveillance cultures in the hospital setting because we have conducted elimination procedures to manage bacterial spread. However, we tried to analyze risk factors for Elizabethkingia acquisition by minimizing the selection bias using a PS-matched study and multivariate analysis.
Even after controlling for potential biases using PS matching analysis, we found mechanical ventilation to be an independent risk factor for the acquisition of Elizabethkingia species. Because Elizabethkingia infection has a high rate of death and is difficult to eliminate, intensive prevention of contamination is needed.
Dr. Choi is a microbiologist at the Department of Laboratory Medicine and Research Institute of Bacterial Resistance, Yonsei University College of Medicine, Seoul, South Korea. His primary research interests include antimicrobial resistance.
References
- Vandamme P, Bernardet J-F, Kersters SK, Holmes B. New perspectives in the classification of the flavobacteria: description of Chryseobacterium gen. nov., Bergeyella gen. nov., and Empedobacter nom. rev. Int J Syst Evol Microbiol. 1994;44:827–31.
- Bloch KC, Nadarajah R, Jacobs R. Chryseobacterium meningosepticum: an emerging pathogen among immunocompromised adults. Report of 6 cases and literature review. Medicine (Baltimore). 1997;76:30–41. DOIPubMedGoogle Scholar
- Li Y, Kawamura Y, Fujiwara N, Naka T, Liu H, Huang X, et al. Chryseobacterium miricola sp. nov., a novel species isolated from condensation water of space station Mir. Syst Appl Microbiol. 2003;26:523–8. DOIPubMedGoogle Scholar
- Green O, Murray P, Gea-Banacloche JC. Sepsis caused by Elizabethkingia miricola successfully treated with tigecycline and levofloxacin. Diagn Microbiol Infect Dis. 2008;62:430–2. DOIPubMedGoogle Scholar
- Kämpfer P, Matthews H, Glaeser SP, Martin K, Lodders N, Faye I. Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int J Syst Evol Microbiol. 2011;61:2670–5. DOIPubMedGoogle Scholar
- Lau SK, Chow WN, Foo CH, Curreem SO, Lo GC, Teng JL, et al. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci Rep. 2016;6:26045. DOIPubMedGoogle Scholar
- Han MS, Kim H, Lee Y, Kim M, Ku NS, Choi JY, et al. Relative prevalence and antimicrobial susceptibility of clinical isolates of Elizabethkingia species based on 16S rRNA gene sequencing. J Clin Microbiol. 2016;55:274–80. DOIPubMedGoogle Scholar
- Teo J, Tan SY, Tay M, Ding Y, Kjelleberg S, Givskov M, et al. First case of E anophelis outbreak in an intensive-care unit. Lancet. 2013;382:855–6. DOIPubMedGoogle Scholar
- Lau SK, Wu AK, Teng JL, Tse H, Curreem SO, Tsui SK, et al. Evidence for Elizabethkingia anophelis transmission from mother to infant, Hong Kong. Emerg Infect Dis. 2015;21:232–41. DOIPubMedGoogle Scholar
- Matyi SA, Hoyt PR, Hosoyama A, Yamazoe A, Fujita N, Gustafson JE. Draft genome sequences of Elizabethkingia meningoseptica. Genome Announc. 2013;1:e00444–13. DOIPubMedGoogle Scholar
- Yum JH, Lee EY, Hur SH, Jeong SH, Lee H, Yong D, et al. Genetic diversity of chromosomal metallo-β-lactamase genes in clinical isolates of Elizabethkingia meningoseptica from Korea. J Microbiol. 2010;48:358–64. DOIPubMedGoogle Scholar
- Chen GX, Zhang R, Zhou HW. Heterogeneity of metallo-β-lactamases in clinical isolates of Chryseobacterium meningosepticum from Hangzhou, China. J Antimicrob Chemother. 2006;57:750–2. DOIPubMedGoogle Scholar
- Moore LS, Owens DS, Jepson A, Turton JF, Ashworth S, Donaldson H, et al. Waterborne Elizabethkingia meningoseptica in adult critical care. Emerg Infect Dis. 2016;22:9–17. DOIPubMedGoogle Scholar
- Balm MN, Salmon S, Jureen R, Teo C, Mahdi R, Seetoh T, et al. Bad design, bad practices, bad bugs: frustrations in controlling an outbreak of Elizabethkingia meningoseptica in intensive care units. J Hosp Infect. 2013;85:134–40. DOIPubMedGoogle Scholar
- Lin PY, Chen HL, Huang CT, Su LH, Chiu CH. Biofilm production, use of intravascular indwelling catheters and inappropriate antimicrobial therapy as predictors of fatality in Chryseobacterium meningosepticum bacteraemia. Int J Antimicrob Agents. 2010;36:436–40. DOIPubMedGoogle Scholar
- Weaver KN, Jones RC, Albright R, Thomas Y, Zambrano CH, Costello M, et al. Acute emergence of Elizabethkingia meningoseptica infection among mechanically ventilated patients in a long-term acute care facility. Infect Control Hosp Epidemiol. 2010;31:54–8. DOIPubMedGoogle Scholar
- Pereira GH, Garcia DO, Abboud CS, Barbosa VL, Silva PS. Nosocomial infections caused by Elizabethkingia meningoseptica: an emergent pathogen. Braz J Infect Dis. 2013;17:606–9. DOIPubMedGoogle Scholar
- Ghafur A, Vidyalakshmi PR, Priyadarshini K, Easow JM, Raj R, Raja T. Elizabethkingia meningoseptica bacteremia in immunocompromised hosts: The first case series from India. South Asian J Cancer. 2013;2:211–5. DOIPubMedGoogle Scholar
- Hsu MS, Liao CH, Huang YT, Liu CY, Yang CJ, Kao KL, et al. Clinical features, antimicrobial susceptibilities, and outcomes of Elizabethkingia meningoseptica (Chryseobacterium meningosepticum) bacteremia at a medical center in Taiwan, 1999-2006. Eur J Clin Microbiol Infect Dis. 2011;30:1271–8. DOIPubMedGoogle Scholar
- Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373–83. DOIPubMedGoogle Scholar
- Tanaka K, Mizusawa K, Saugstad ES. A revision of the adult and larval mosquitoes of Japan (including the Ryukyu Archipelago and the Ogasawara Islands) and Korea (Diptera: Culicidae): San Francisco: US Army Medical Lab Pacific; 1979.
- Lindh JM, Terenius O, Faye I. 16S rRNA gene-based identification of midgut bacteria from field-caught Anopheles gambiae sensu lato and A. funestus mosquitoes reveals new species related to known insect symbionts. Appl Environ Microbiol. 2005;71:7217–23. DOIPubMedGoogle Scholar
- Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. CDC definitions for nosocomial infections, 1988. Am J Infect Control. 1988;16:128–40. DOIPubMedGoogle Scholar
- Brady OJ, Smith DL, Scott TW, Hay SI. Dengue disease outbreak definitions are implicitly variable. Epidemics. 2015;11:92–102. DOIPubMedGoogle Scholar
- Wagner MM, Tsui FC, Espino JU, Dato VM, Sittig DF, Caruana RA, et al. The emerging science of very early detection of disease outbreaks. J Public Health Manag Pract. 2001;7:51–9. DOIPubMedGoogle Scholar
- Wyss R, Girman CJ, LoCasale RJ, Brookhart AM, Stürmer T. Variable selection for propensity score models when estimating treatment effects on multiple outcomes: a simulation study. Pharmacoepidemiol Drug Saf. 2013;22:77–85. DOIPubMedGoogle Scholar
- Hill J. Discussion of research using propensity-score matching: comments on ‘A critical appraisal of propensity-score matching in the medical literature between 1996 and 2003’ by Peter Austin, Statistics in Medicine. Stat Med. 2008;27:2055–61, discussion 2066–9. DOIPubMedGoogle Scholar
- Perrin A, Larsonneur E, Nicholson AC, Edwards DJ, Gundlach KM, Whitney AM, et al. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat Commun. 2017;8:15483. DOIPubMedGoogle Scholar
- Jean SS, Lee WS, Chen FL, Ou TY, Hsueh PR. Elizabethkingia meningoseptica: an important emerging pathogen causing healthcare-associated infections. J Hosp Infect. 2014;86:244–9. DOIPubMedGoogle Scholar
- Lin YT, Chiu CH, Chan YJ, Lin ML, Yu KW, Wang FD, et al. Clinical and microbiological analysis of Elizabethkingia meningoseptica bacteremia in adult patients in Taiwan. Scand J Infect Dis. 2009;41:628–34. DOIPubMedGoogle Scholar
- Kanamori H, Weber DJ, Rutala WA. Healthcare outbreaks associated with a water reservoir and infection prevention strategies. Clin Infect Dis. 2016;62:1423–35. DOIPubMedGoogle Scholar
- Ceyhan M, Yildirim I, Tekeli A, Yurdakok M, Us E, Altun B, et al. A Chryseobacterium meningosepticum outbreak observed in 3 clusters involving both neonatal and non-neonatal pediatric patients. Am J Infect Control. 2008;36:453–7. DOIPubMedGoogle Scholar
- Sader HS, Jones RN. Antimicrobial susceptibility of uncommonly isolated non-enteric Gram-negative bacilli. Int J Antimicrob Agents. 2005;25:95–109. DOIPubMedGoogle Scholar
- Kirby JT, Sader HS, Walsh TR, Jones RN. Antimicrobial susceptibility and epidemiology of a worldwide collection of Chryseobacterium spp: report from the SENTRY Antimicrobial Surveillance Program (1997-2001). J Clin Microbiol. 2004;42:445–8. DOIPubMedGoogle Scholar
- Price E, Hoffman P, Weaver G, Gilks J, Jones M, O’Brien V, et al. Difficulty with decontaminating dummies (pacifiers, soothers or comforters) for infants in hospital. J Hosp Infect. 2017;97:316. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: November 27, 2018
1These authors contributed equally to this article.
Table of Contents – Volume 25, Number 1—January 2019
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Jun Yong Choi, Department of Internal Medicine and AIDS Research Institute, Severance Hospital, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, South Korea; ; or Kyungwon Lee, Department of Laboratory Medicine, Severance Hospital, Research Institute of Bacterial Resistance, Yonsei University College of Medicine, 50-1 Yonsei-ro, Seodaemun-gu, Seoul 03722, South Korea
Top