Volume 25, Number 1—January 2019
Dispatch
Puumala Hantavirus Genotypes in Humans, France, 2012–2016
Figure 1
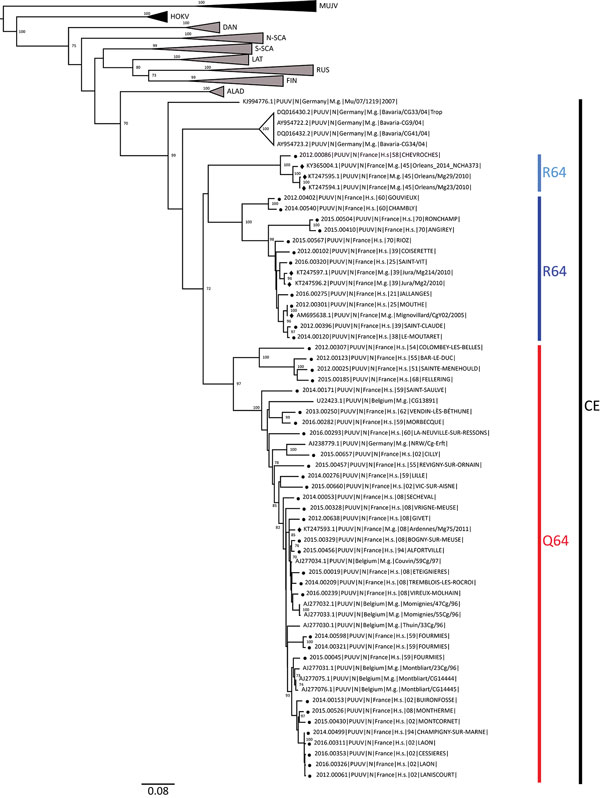
Figure 1. Phylogenetic tree constructed using the maximum-likelihood approach based on the complete small-segment RNA nucleotide coding sequences of representative Puumala virus (PUUV) strains detected in human cases in France, 2012–2016 (circles), and on those published and representative of PUUV strains detected in Europe. Diamonds indicate sequences of strains detected in rodents as reported elsewhere (7,8). Bootstrap percentages >70% (from 1,000 resamplings) are indicated at each node; GenBank accession numbers are indicated for reference strains. Scale bar indicates nucleotide substitutions per site. ALAD, Alpe-Adrian lineage; CE, Central European lineage; DAN, Danish lineage; FIN, Finnish lineage; LAT, Latvian lineage; N-SCA, north-Scandinavian lineage; S-SCA; south-Scandinavian lineage; RUS, Russian lineage; MUJV, Muju virus; HOKV, Hokkaido virus.
References
- Vaheri A, Henttonen H, Voutilainen L, Mustonen J, Sironen T, Vapalahti O. Hantavirus infections in Europe and their impact on public health. Rev Med Virol. 2013;23:35–49. DOIPubMedGoogle Scholar
- International Committee on Taxonomy of Viruses (ICTV). 2016.023a-cM Hantavirus approved proposal. 2017 March 19. [cited 2018 Feb 9]. https://talk.ictvonline.org/ICTV/proposals/2016.023a-cM.A.v2.Hantavirus_sprev.pdf
- Sironen T, Vaheri A, Plyusnin A. Molecular evolution of Puumala hantavirus. J Virol. 2001;75:11803–10. DOIPubMedGoogle Scholar
- Razzauti M, Plyusnina A, Niemimaa J, Henttonen H, Plyusnin A. Co-circulation of two Puumala hantavirus lineages in Latvia: a Russian lineage described previously and a novel Latvian lineage. J Med Virol. 2012;84:314–8. DOIPubMedGoogle Scholar
- Evander M, Eriksson I, Pettersson L, Juto P, Ahlm C, Olsson GE, et al. Puumala hantavirus viremia diagnosed by real-time reverse transcriptase PCR using samples from patients with hemorrhagic fever and renal syndrome. J Clin Microbiol. 2007;45:2491–7. DOIPubMedGoogle Scholar
- Reynes JM, Carli D, Renaudin B, Fizet A, Bour JB, Brodard V, et al. Surveillance of human hantavirus infections in metropolitan France 2012–2016. [in French]. Bull Epidemiol Hebd (Paris). 2017;23:492–9.
- Plyusnina A, Deter J, Charbonnel N, Cosson JF, Plyusnin A. Puumala and Tula hantaviruses in France. Virus Res. 2007;129:58–63. DOIPubMedGoogle Scholar
- Castel G, Couteaudier M, Sauvage F, Pons JB, Murri S, Plyusnina A, et al. Complete genome and phylogeny of Puumala hantavirus isolates circulating in France. Viruses. 2015;7:5476–88. DOIPubMedGoogle Scholar
- Kramski M, Meisel H, Klempa B, Krüger DH, Pauli G, Nitsche A. Detection and typing of human pathogenic hantaviruses by real-time reverse transcription-PCR and pyrosequencing. Clin Chem. 2007;53:1899–905. DOIPubMedGoogle Scholar
- Bowen MD, Gelbmann W, Ksiazek TG, Nichol ST, Nowotny N. Puumala virus and two genetic variants of Tula virus are present in Austrian rodents. J Med Virol. 1997;53:174–81. DOIPubMedGoogle Scholar
- Aitichou M, Saleh SS, McElroy AK, Schmaljohn C, Ibrahim MS. Identification of Dobrava, Hantaan, Seoul, and Puumala viruses by one-step real-time RT-PCR. J Virol Methods. 2005;124:21–6. DOIPubMedGoogle Scholar
- Garin D, Peyrefitte C, Crance JM, Le Faou A, Jouan A, Bouloy M. Highly sensitive Taqman PCR detection of Puumala hantavirus. Microbes Infect. 2001;3:739–45. DOIPubMedGoogle Scholar
- Korva M, Saksida A, Kejžar N, Schmaljohn C, Avšič-Županc T. Viral load and immune response dynamics in patients with haemorrhagic fever with renal syndrome. Clin Microbiol Infect. 2013;19:E358–66. DOIPubMedGoogle Scholar
- Lagerqvist N, Hagström Å, Lundahl M, Nilsson E, Juremalm M, Larsson I, et al. Molecular diagnosis of hemorrhagic fever with renal syndrome caused by Puumala virus. J Clin Microbiol. 2016;54:1335–9. DOIPubMedGoogle Scholar
- Razzauti M, Plyusnina A, Sironen T, Henttonen H, Plyusnin A. Analysis of Puumala hantavirus in a bank vole population in northern Finland: evidence for co-circulation of two genetic lineages and frequent reassortment between strains. J Gen Virol. 2009;90:1923–31. DOIPubMedGoogle Scholar
1Current affiliation: Institut Pasteur, Paris, France.
2Current affiliation: Laboratoire P4 INSERM Jean Mérieux, Lyon, France.