Volume 25, Number 1—January 2019
Research Letter
Phylogeographic Analysis of African Swine Fever Virus, Western Europe, 2018
Figure
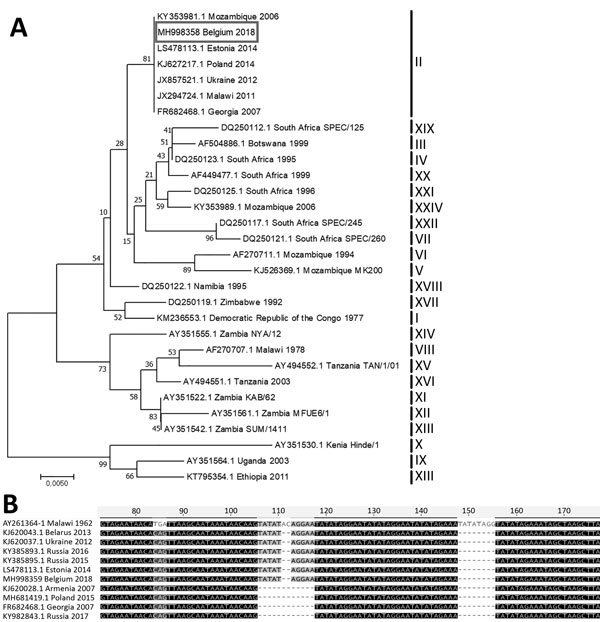
Figure. A) Evolutionary relationships of representative strains of African swine fever virus based on the neighbor-joining phylogeny of the partial p72 gene sequences. The phylogenetic analysis was performed using MEGA7 (http://www.megasoftware.net) and the Kimura 2-parameter substitution model, as determined by a model selection analysis. Bootstrap values (>70%, based on 500 replicates) for each node are given. GenBank accession numbers, country, and year of collection are indicated for each strain; for strains for which the year of collection is not known, the strain name is indicated. Corresponding genotypes are labeled I–XXIV. Box indicates the African swine fever sequence from Belgium generated during this study. Scale bar indicates nucleotide substitutions per site. B) Nucleotide sequence alignment of the partial intergenic region between the I73R and I329L genes from representative African swine fever virus strains. GenBank accession numbers, country, and year of collection are indicated.