Volume 25, Number 12—December 2019
Research
Genomic Analysis of Fluoroquinolone- and Tetracycline-Resistant Campylobacter jejuni Sequence Type 6964 in Humans and Poultry, New Zealand, 2014–2016
Figure 2
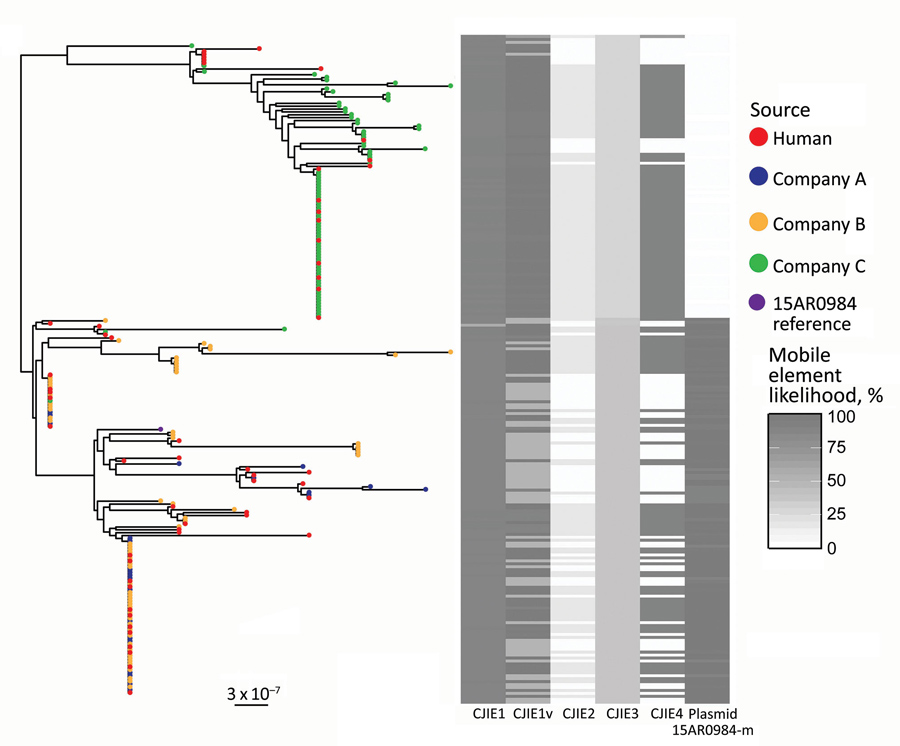
Figure 2. Population structure of 227 sequence type 6964 Campylobacter jejuni isolates from humans and poultry, New Zealand, 2014–2016. The tree is the inferred midpoint rooted phylogeny of the isolates, including the reference 15AR0984 genome. The tips are colored by source of the C. jejuni isolate. The heatmap indicates the likelihoods of the presence of mobile elements including CJIE1 variant (cjie1_15AR0984), CJIEs 1–4, and the plasmid 15AR0984-m. Dark shading on the heatmap indicates 100% likelihood; white indicates absence. Scale bar indicates nucleotide substitutions per site.
References
- Kaakoush NO, Castaño-Rodríguez N, Mitchell HM, Man SM. Global epidemiology of Campylobacter infection. Clin Microbiol Rev. 2015;28:687–720. DOIPubMedGoogle Scholar
- Sproston EL, Wimalarathna HML, Sheppard SK. Trends in fluoroquinolone resistance in Campylobacter. Microb Genom. 2018;4.
- The Institute of Environmental Science and Research Ltd. Notifiable diseases in New Zealand: annual report 2017 [cited 2019 Oct 9]. https://surv.esr.cri.nz/surveillance/annual_surveillance.php
- Sears A, Baker MG, Wilson N, Marshall J, Muellner P, Campbell DM, et al. Marked campylobacteriosis decline after interventions aimed at poultry, New Zealand. Emerg Infect Dis. 2011;17:1007–15. DOIPubMedGoogle Scholar
- Müllner P, Collins-Emerson JM, Midwinter AC, Carter P, Spencer SE, van der Logt P, et al. Molecular epidemiology of Campylobacter jejuni in a geographically isolated country with a uniquely structured poultry industry. Appl Environ Microbiol. 2010;76:2145–54. DOIPubMedGoogle Scholar
- McTavish SM, Pope CE, Nicol C, Sexton K, French N, Carter PE. Wide geographical distribution of internationally rare Campylobacter clones within New Zealand. Epidemiol Infect. 2008;136:1244–52. DOIPubMedGoogle Scholar
- Mullner P, Spencer SEF, Wilson DJ, Jones G, Noble AD, Midwinter AC, et al. Assigning the source of human campylobacteriosis in New Zealand: a comparative genetic and epidemiological approach. Infect Genet Evol. 2009;9:1311–9. DOIPubMedGoogle Scholar
- Sheppard SK, Cheng L, Méric G, de Haan CP, Llarena A-K, Marttinen P, et al. Cryptic ecology among host generalist Campylobacter jejuni in domestic animals. Mol Ecol. 2014;23:2442–51. DOIPubMedGoogle Scholar
- Bolwell CF, Gilpin BJ, Campbell D, French NP. Evaluation of the representativeness of a sentinel surveillance site for campylobacteriosis. Epidemiol Infect. 2015;143:1990–2002. DOIPubMedGoogle Scholar
- Williamson D, Dyet K, Heffernan H. Antimicrobial resistance in human isolates of Campylobacter jejuni, 2015 [cited 2019 Oct 9]. https://surv.esr.cri.nz/PDF_surveillance/Antimicrobial/CAMPY/CampyFQRfinalreport2015.pdf
- Nohra A, Grinberg A, Midwinter AC, Marshall JC, Collins-Emerson JM, French NP. Molecular epidemiology of Campylobacter coli strains isolated from different sources in New Zealand between 2005 and 2014. Appl Environ Microbiol. 2016;82:4363–70. DOIPubMedGoogle Scholar
- Wang G, Clark CG, Taylor TM, Pucknell C, Barton C, Price L, et al. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J Clin Microbiol. 2002;40:4744–7. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria, 3rd edition (M45). Wayne (PA): The Institute; 2016.
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 5.0. 2015 [cited 2018 Oct 10]. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_5.0_Breakpoint_Table_01.pdf
- Dingle KE, McCarthy ND, Cody AJ, Peto TE, Maiden MC. Extended sequence typing of Campylobacter spp., United Kingdom. Emerg Infect Dis. 2008;14:1620–2. DOIPubMedGoogle Scholar
- Baines SL, Howden BP, Heffernan H, Stinear TP, Carter GP, Seemann T, et al. Rapid emergence and evolution of Staphylococcus aureus clones harboring fusC-containing staphylococcal cassette chromosome elements. Antimicrob Agents Chemother. 2016;60:2359–65. DOIPubMedGoogle Scholar
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Zhang J, Xiong Y, Rogers L, Carter GP, French N. Genome-by-genome approach for fast bacterial genealogical relationship evaluation. Bioinformatics. 2018;34:3025–7. DOIPubMedGoogle Scholar
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–67. DOIPubMedGoogle Scholar
- Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:
e15 . DOIPubMedGoogle Scholar - Cody AJ, Bray JE, Jolley KA, McCarthy ND, Maiden MCJ. Core genome multilocus sequence typing scheme for stable, comparative analyses of Campylobacter jejuni and C. coli human disease isolates. J Clin Microbiol. 2017;55:2086–97. DOIPubMedGoogle Scholar
- Page AJ, Taylor B, Delaney AJ, Soares J, Seemann T, Keane JA, et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom. 2016;2:
e000056 . DOIPubMedGoogle Scholar - Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. DOIPubMedGoogle Scholar
- Minh BQ, Nguyen MA, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–95. DOIPubMedGoogle Scholar
- Yu G, Lam TT, Zhu H, Guan Y. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol Biol Evol. 2018;35:3041–3. DOIPubMedGoogle Scholar
- Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, Harris SR. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics. 2017. [Epub ahead of print].
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. DOIPubMedGoogle Scholar
- Llarena A-K, Zhang J, Vehkala M, Välimäki N, Hakkinen M, Hänninen M-L, et al. Monomorphic genotypes within a generalist lineage of Campylobacter jejuni show signs of global dispersion. Microb Genom. 2016;2:
e000088 . DOIPubMedGoogle Scholar - Parker CT, Quiñones B, Miller WG, Horn ST, Mandrell RE. Comparative genomic analysis of Campylobacter jejuni strains reveals diversity due to genomic elements similar to those present in C. jejuni strain RM1221. J Clin Microbiol. 2006;44:4125–35. DOIPubMedGoogle Scholar
- Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–403. DOIPubMedGoogle Scholar
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. DOIPubMedGoogle Scholar
- Poly F, Read TD, Chen YH, Monteiro MA, Serichantalergs O, Pootong P, et al. Characterization of two Campylobacter jejuni strains for use in volunteer experimental-infection studies. Infect Immun. 2008;76:5655–67. DOIPubMedGoogle Scholar
- Wang Y, Huang WM, Taylor DE. Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob Agents Chemother. 1993;37:457–63. DOIPubMedGoogle Scholar
- Marasini D, Fakhr MK. Whole-genome sequencing of a Campylobacter jejuni strain isolated from retail chicken meat reveals the presence of a megaplasmid with Mu-like prophage and multidrug resistance genes. Genome Announc. 2016;4:e00460–16. DOIPubMedGoogle Scholar
- Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species. PLoS Biol. 2005;3:
e15 . DOIPubMedGoogle Scholar - The Institute of Environmental Science and Research Ltd. General antimicrobial susceptibility data collected from hospital and community laboratories [cited 2018 Aug 1]. https://surv.esr.cri.nz/antimicrobial/general_antimicrobial_susceptibility.php
- Pleydell EJ, Rogers L, Kwan E, French NP. Low levels of antibacterial drug resistance expressed by Gram-negative bacteria isolated from poultry carcasses in New Zealand. N Z Vet J. 2010;58:229–36. DOIPubMedGoogle Scholar
- Heffernen H, Wong T, Lindsay J, Bowen B, Woodhouse R. A baseline survey of antimicrobial resistance in bacteria from selected New Zealand foods, 2009–2010 [cited 2019 Oct 9]. https://www.mpi.govt.nz/dmsdocument/21464-a-baseline-survey-of-antimicrobial-resistance-in-bacteria-from-selected-new-zealand-foods-2009-2010
- New Zealand Food Safety. Antibiotic sales analysis 2014–2016. Technical paper no. 2018/08 [cited 2019 Oct 9]. https://www.fisheries.govt.nz/dmsdocument/31920/direct
- Ministry for Primary Industries. 2011–2014 Antibiotic Sales Analysis. MPI technical paper no. 2016/65 [cited 2019 Oct 9]. https://www.mpi.govt.nz/dmsdocument/14497-2011-2014-antibiotic-sales-analysis
- Huang JY, Henao OL, Griffin PM, Vugia DJ, Cronquist AB, Hurd S, et al. Infection with pathogens transmitted commonly through food and the effect of increasing use of culture-independent diagnostic tests on surveillance—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2012–2015. MMWR Morb Mortal Wkly Rep. 2016;65:368–71. DOIPubMedGoogle Scholar
- May FJ, Stafford RJ, Carroll H, Robson JM, Vohra R, Nimmo GR, et al. The effects of culture independent diagnostic testing on the diagnosis and reporting of enteric bacterial pathogens in Queensland, 2010 to 2014. Commun Dis Intell Q Rep. 2017;41:E223–30.PubMedGoogle Scholar
- Whitehouse CA, Young S, Li C, Hsu CH, Martin G, Zhao S. Use of whole-genome sequencing for Campylobacter surveillance from NARMS retail poultry in the United States in 2015. Food Microbiol. 2018;73:122–8. DOIPubMedGoogle Scholar
- Clark CG, Grant CC, Pollari F, Marshall B, Moses J, Tracz DM, et al. Effects of the Campylobacter jejuni CJIE1 prophage homologs on adherence and invasion in culture, patient symptoms, and source of infection. BMC Microbiol. 2012;12:269. DOIPubMedGoogle Scholar
- Clark CG, Chong PM, McCorrister SJ, Simon P, Walker M, Lee DM, et al. The CJIE1 prophage of Campylobacter jejuni affects protein expression in growth media with and without bile salts. BMC Microbiol. 2014;14:70. DOIPubMedGoogle Scholar
- Gaasbeek EJ, Wagenaar JA, Guilhabert MR, van Putten JP, Parker CT, van der Wal FJ. Nucleases encoded by the integrated elements CJIE2 and CJIE4 inhibit natural transformation of Campylobacter jejuni. J Bacteriol. 2010;192:936–41. DOIPubMedGoogle Scholar
- Gaasbeek EJ, Wagenaar JA, Guilhabert MR, Wösten MM, van Putten JP, van der Graaf-van Bloois L, et al. A DNase encoded by integrated element CJIE1 inhibits natural transformation of Campylobacter jejuni. J Bacteriol. 2009;191:2296–306. DOIPubMedGoogle Scholar
Page created: November 18, 2019
Page updated: November 18, 2019
Page reviewed: November 18, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.