Volume 25, Number 3—March 2019
Dispatch
Whole-Genome Sequencing of Drug-Resistant Mycobacterium tuberculosis Strains, Tunisia, 2012–2016
Figure 2
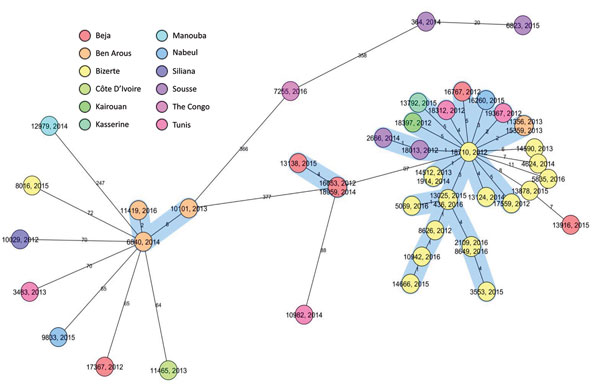
Figure 2. Core-genome multilocus sequence typing–based minimum spanning tree for strains identified during study of drug-resistant Mycobacterium tuberculosis, Tunisia, 2012–2016. Ridom SeqSphere+ minimum spanning tree for 46 samples based on 2,891 columns, pairwise ignoring missing values, logarithmic scale. Cluster distance threshold = 5 alleles. Colors indicate regions and countries of patient origin.
Page created: February 19, 2019
Page updated: February 19, 2019
Page reviewed: February 19, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.