Volume 25, Number 4—April 2019
Dispatch
Streptococcus agalactiae Sequence Type 283 in Farmed Fish, Brazil
Cite This Article
Citation for Media
Abstract
In 2016 and 2017, we characterized outbreaks caused by Streptococcus agalactiae serotype III sequence type (ST) 283 in Nile tilapia farms in Brazil. Whole-genome multilocus sequence typing clustered the fish isolates together with the zoonotic ST283 and other STs related to cases in humans, frogs, dogs, cattle, and dolphins.
Streptococcus agalactiae (group B Streptococcus [GBS]) is a major etiologic agent of diseases in humans and animals (1). Episodes of bacteremia and meningitis in humans associated with raw fish consumption were reported in Singapore (2,3). Two case–control studies determined that GBS serotype III sequence type (ST) 283 was associated with disease in 9 (2) and 19 (3) patients in that country at different times during 2015. This specific ST has been identified as a zoonotic agent to humans and already has been detected in freshwater fish dishes from food stalls in Singapore (1); consumption of such fish dishes led to increased cases during that year. In farmed fish, GBS serotype III ST283 was detected in diseased tilapia (Oreochromis sp.) in Thailand (4).
In Brazil, fish-pathogenic GBS has been isolated mainly from farmed Nile tilapia (Oreochromis niloticus) since the 2000s. The predominant serotype in Brazil is serotype Ib from ST260, ST927, and a nontypeable ST; all these STs are reported as fish-adapted pathogens (5). Sporadic detection of serotype Ia ST103 also was described in Brazil (6). However, since 2016, a new serotype, serotype III, has emerged in the country. This serotype has been detected in different Nile tilapia farms in the northeastern region (7). The genetic diversity and ST of these isolates have not yet been identified.
Our objective was to evaluate, by molecular and genomic approaches, the GBS serotype III isolates from outbreaks in Nile tilapia farms in Brazil. We also aimed to study the genetic relationship between isolates from farmed fish in Brazil, from foodborne isolates from humans in Asia, and from cases in fish.
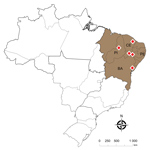
During July 2016–June of 2017, we followed 6 outbreaks that led to Nile tilapia deaths at commercial farms in 4 Brazilian states: Piauí, Pernambuco, Ceará, and Bahia (Figure 1). Disease outbreaks with high death rates (daily rates >0.2% and total death rates in the herds of 25%–35%) in fish vaccinated with a commercial inactivated vaccine against GBS serotype Ib were reported. Diseased fish showed signs of lethargy, melanosis, and exophthalmia. We sampled 71 diseased fish and transported them on ice to the laboratory (Table), where we immediately performed bacteriologic analyses. Fish tissue sampling and the culture conditions of the analyses were as described previously (8). The isolates obtained from 64 fish were identified up to the species level by matrix-assisted laser desorption ionization time-of-flight mass spectrometry analysis, using a previously published method (9). We confirmed GBS as the principal pathogen associated with the deaths during all the outbreaks (Table).
We performed capsular polysaccharide typing for all GBS isolates using a previously published multiplex PCR (10). In addition, we conducted sequencing of housekeeping genes for multilocus sequence typing (MLST) analysis (11). According to the GBS MLST database (http://pubmlst.org/sagalactiae), 59 isolates belonged to serotype III ST283 and 3 to serotype Ib ST260.
For selected GBS isolates from each farm, we performed a PCR-based fingerprinting technique (ERIC-PCR [enterobacterial repetitive intergenic consensus PCR]) described by Costa et al. (12) to evaluate genetic diversity in comparison with MLST results. We observed nondistinguishable banding patterns between all GBS serotype III ST283 isolates, regardless of their geographic origin (data not shown), suggesting the possibility of a genetically close relation.
To determine the genomic relationship between GBS serotype III ST283 isolates from fish in Brazil and Asia and foodborne cases in humans, we conducted whole-genome sequencing and whole-genome MLST (wgMLST) analyses. In brief, 8 isolates (SA01AQUAVET, SA06AQUAVET, SA12AQUAVET, SA22AQUAVET, SA90AQUAVET, SA98AQUAVET, SApx2AQUAVET, and SApx7AQUAVET), representing each outbreak case, and 3 isolates from outbreak 2, because the isolates of outbreak 2 were obtained from 3 different farms, were selected (Appendix).
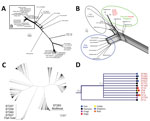
In the wgMLST analysis, GBS serotype III ST283 isolates formed 1 phylogenomic related group (Figure 2, panel A). We compared 3,539 loci of the analyzed GBS isolates based on pairwise comparisons of the numbers of homologous loci with distinct allele sequences and found that all ST283 isolates (from fish and humans) displayed up to 505 distinct allele sequences. When we compared isolates from Brazil with fish strains from Asia, we observed variations ranging from 136 to 505 (3.84%–14.26%) (Figure 2, panel B) in loci. Among isolates from humans and fish in Asia, the variations were 31–240 (0.87%–6.78%), whereas for isolates from humans in Asia compared with from fish in Brazil, the variations were 272–415 (7.68%–11.72%). GBS ST283 from Brazil showed loci variations ranging from 71 to 256 between each other. These levels of local diversities within an ST were reported in a previous study that evaluated distinct STs obtained from fish in Brazil (5).
Finally, we conducted a phylogenetic analysis with MLST loci concatenated (Figure 2, panels C, D). The 7 loci (i.e., adhP, pheS, atr, glnA, sdhA, glcK, and tkt) of each of 1,193 STs, available in GBS MLST database) were concatenated and aligned using CLC Genomics Workbench (QIAGEN, https://www.qiagen.com). From the alignment results, we generated a phylogenetic tree in the CLC Genomics Workbench using the UPGMA (unweighted pair group method with arithmetic mean) construction method, Jukes-Cantor as the nucleotide distance measure, and 1,000 replicates on bootstrap analysis. ST283 formed a consistent (bootstrap 90%) clade with other STs with different hosts other than fish (Figure 2, panel D), such as humans, fish, and frogs, including some with multihost range, among them humans, fish, cattle, dolphins, dogs, and frogs.
We described the emergence of GBS ST (GBS serotype III ST283) associated with the Nile tilapia infection in Brazil. ST283 had been previously detected in fish (with and without clinical disease) only in Southeast Asia countries (1,4). This report in Brazil indicates possible spread of this pathogen genotype around the world. Based on import records of live Nile tilapia from Singapore to Brazil in 2014 and our analyses (ERIC-PCR, MLST, and wgMLST), it is possible that this genotype was introduced into the country with the recently imported fish.
GBS serotype III ST283 caused severe foodborne outbreaks in Singapore in 2015, and these outbreaks have been linked to raw fish consumption (2), indicating a zoonotic lineage for this serotype. Our results demonstrate that GBS serotype III ST283 isolates from Brazil clustered in the same network branch with GBS isolates from Asia, from fish and human hosts; however, we observed genomic diversity that depicted a clonal expansion of this genotype. The genomic diversity among GBS isolates from human and fish hosts was also observed in a previous study (1), which further reinforces the concept of a lineage that is adapted to >1 host. This previous study on GBS suggested that ST283 might not be pathogenic for fish (1). However, a previous study (4) found that it was associated with disease in fish. After evaluating isolates of tilapia clinical disease cases, we confirmed that this ST283 is associated with multiple outbreaks in commercial tilapia farms. Thus, it demonstrates the clear pathogenic behavior of this ST to fish. Moreover, genomic analysis indicates it belongs to a cluster with other STs from multiple susceptible hosts (Figure 2, panel D), such as the ST12, which was isolated from humans, dogs, dolphins, and rats, corroborating the suggestion that the member of this clade affects multiple host species. ST283 was only isolated from diseased tilapia, which cannot be used for human consumption. Future longitudinal noncasuistic study is needed to identify the occurrence and prevalence of GBS infection by the ST283 lineage also in asymptomatic fish before and after the outbreaks. In this context, asymptomatic tilapia carriers might represent a serious public health concern.
Dr. Leal is a veterinarian and adjunct professor at the Federal University of Minas Gerais, Brazil. His primary research interest is infectious diseases in aquatic animals, especially bacterial diseases, and the immunology of these animals.
Acknowledgment
Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) provided student fellowships, and the Ministry of Agriculture, Livestock and Food Supply financially supported this project.
References
- Chau ML, Chen SL, Yap M, Hartantyo SHP, Chiew PKT, Fernandez CJ, et al. Group B streptococcus infections caused by improper sourcing and handling of fish for raw consumption, Singapore, 2015–2016. Emerg Infect Dis. 2017;23:2002. DOIPubMedGoogle Scholar
- Rajendram P, Mar Kyaw W, Leo YS, Ho H, Chen WK, Lin R, et al. Group B streptococcus sequence type 283 disease linked to consumption of raw fish, Singapore. Emerg Infect Dis. 2016;22:1974–7. DOIPubMedGoogle Scholar
- Tan S, Lin Y, Foo K, Koh HF, Tow C, Zhang Y, et al. Group B streptococcus serotype III sequence type 283 bacteremia associated with consumption of raw fish, Singapore. Emerg Infect Dis. 2016;22:1970–3. DOIPubMedGoogle Scholar
- Delannoy CM, Crumlish M, Fontaine MC, Pollock J, Foster G, Dagleish MP, et al. Human Streptococcus agalactiae strains in aquatic mammals and fish. BMC Microbiol. 2013;13:41. DOIPubMedGoogle Scholar
- Barony GM, Tavares GC, Pereira FL, Carvalho AF, Dorella FA, Leal CAG, et al. Large-scale genomic analyses reveal the population structure and evolutionary trends of Streptococcus agalactiae strains in Brazilian fish farms. Sci Rep. 2017;7:13538. DOIPubMedGoogle Scholar
- Godoy DT, Carvalho-Castro GA, Leal CAG, Pereira UP, Leite RC, Figueiredo HCP. Genetic diversity and new genotyping scheme for fish pathogenic Streptococcus agalactiae. Lett Appl Microbiol. 2013;57:476–83. DOIPubMedGoogle Scholar
- Chideroli RT, Amoroso N, Mainardi RM, Suphoronski SA, de Padua SB, Alfieri AF, et al. Emergence of a new multidrug-resistant and highly virulent serotype of Streptococcus agalactiae in fish farms from Brazil. Aquaculture. 2017;479:45–51. DOIGoogle Scholar
- Mian GF, Godoy DT, Leal CAG, Yuhara TY, Costa GM, Figueiredo HCP. Aspects of the natural history and virulence of S. agalactiae infection in Nile tilapia. Vet Microbiol. 2009;136:180–3. DOIPubMedGoogle Scholar
- Assis GBN, Pereira FL, Zegarra AU, Tavares GC, Leal CAG, Figueiredo HCP. Use of MALDI-TOF mass spectrometry for the fast identification of gram-positive fish pathogens. Front Microbiol. 2017;8:1492. DOIPubMedGoogle Scholar
- Poyart C, Tazi A, Réglier-Poupet H, Billoët A, Tavares N, Raymond J, et al. Multiplex PCR assay for rapid and accurate capsular typing of group B streptococci. J Clin Microbiol. 2007;45:1985–8. DOIPubMedGoogle Scholar
- Jones N, Bohnsack JF, Takahashi S, Oliver KA, Chan M-S, Kunst F, et al. Multilocus sequence typing system for group B streptococcus. J Clin Microbiol. 2003;41:2530–6. DOIPubMedGoogle Scholar
- Costa FAA, Leal CAG, Leite RC, Figueiredo HCP. Genotyping of Streptococcus dysgalactiae strains isolated from Nile tilapia, Oreochromis niloticus (L.). J Fish Dis. 2014;37:463–9. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: March 13, 2019
Table of Contents – Volume 25, Number 4—April 2019
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Carlos A.G. Leal, Veterinary School, Department of Preventive Veterinary Medicine, Federal University of Minas Gerais, Av. Antônio Carlos 6627, Pampulha 31270-901, Belo Horizonte, Minas Gerais, Brazil
Top