Volume 25, Number 5—May 2019
Dispatch
Lassa and Crimean-Congo Hemorrhagic Fever Viruses, Mali
Figure 2
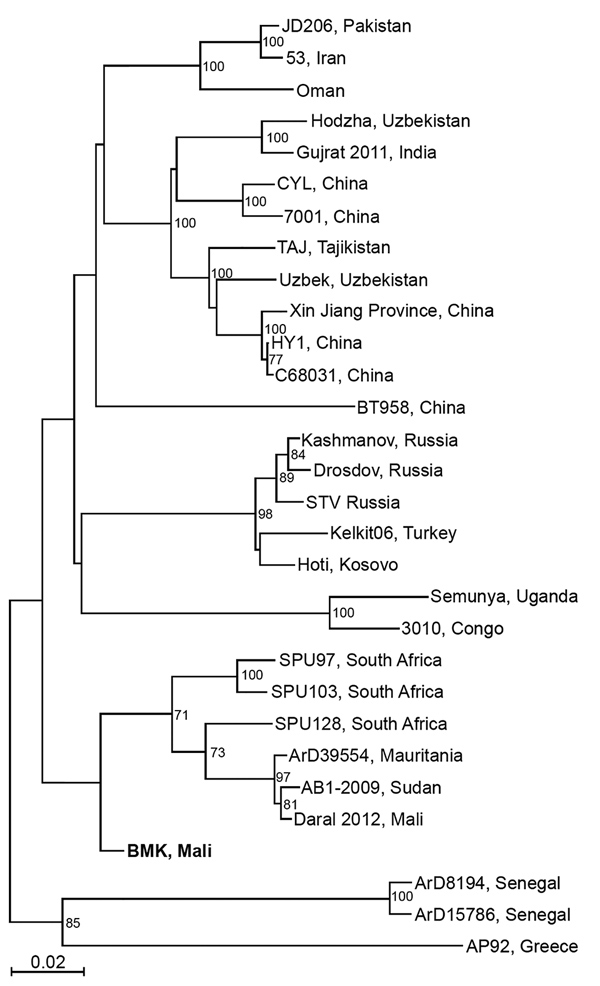
Figure 2. Phylogenetic analysis of representative Crimean-Congo hemorrhagic fever virus (CCHFV) isolates identified in Mali in 2017 (bold) and reference isolates. The tree was constructed by using full-length sequences of the small RNA segment and the neighbor-joining method with bootstrapping to 10,000 iterations. Partial sequences were compared by using the pairwise deletion method. The tree is drawn to scale. Evolutionary analyses were constructed in MEGA7 (https://www.megasoftware.net). Scale bar indicates nucleotide substitutions per site.
1These authors contributed equally to this article.
2These authors were co–principal investigators.
Page created: April 17, 2019
Page updated: April 17, 2019
Page reviewed: April 17, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.