Volume 25, Number 8—August 2019
Research Letter
Multidrug-Resistant Klebsiella pneumoniae ST307 in Traveler Returning from Puerto Rico to Dominican Republic
Figure
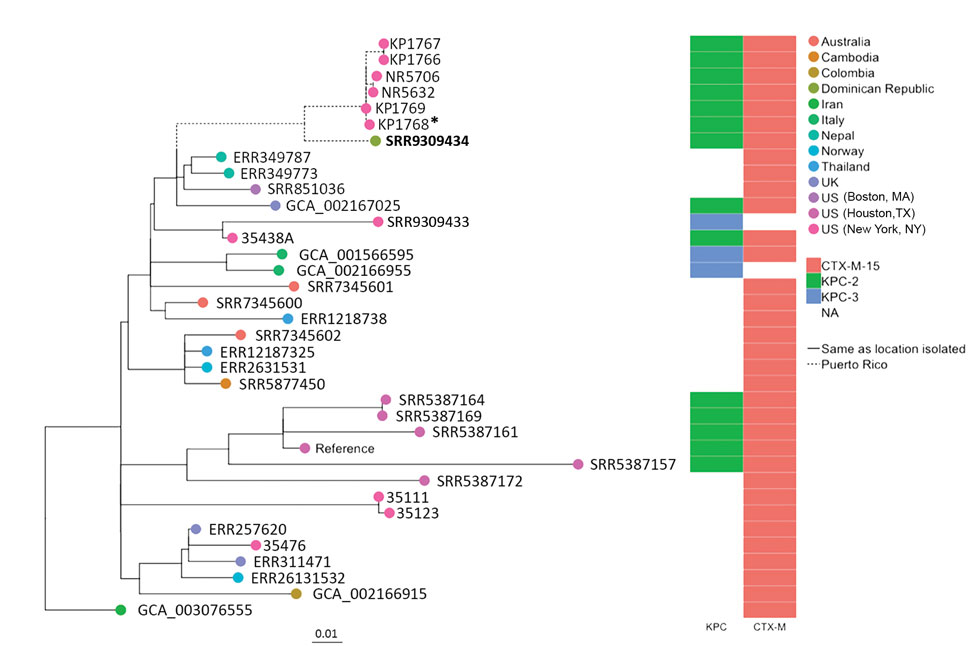
Figure. Maximum-likelihood phylogenetic tree of geographically diverse Klebsiella pneumoniae sequence type 307 isolates based on 860 concatenated single-nucleotide polymorphisms, extracted from an alignment length of 5,248,133 bp. Bold indicates isolate from a traveler from Puerto Rico to the Dominican Republic (this study). Asterisk (*) indicates an isolate recovered from a patient admitted to a hospital in Puerto Rico during the same year as the case-patient in this study (4). bla gene types (KPC, CTX-M) are indicated. Scale bar indicates nucleotide substitutions per site. NA, not applicable.
References
- US Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2013. 2013 Apr [cited 2017 Oct 10]. http://www.cdc.gov/drugresistance/threat-report-2013/index.html
- Zarkotou O, Pournaras S, Tselioti P, Dragoumanos V, Pitiriga V, Ranellou K, et al. Predictors of mortality in patients with bloodstream infections caused by KPC-producing Klebsiella pneumoniae and impact of appropriate antimicrobial treatment. Clin Microbiol Infect. 2011;17:1798–803. DOIPubMedGoogle Scholar
- Long SW, Olsen RJ, Eagar TN, Beres SB, Zhao P, Davis JJ, et al. Population genomic analysis of 1,777 extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates, Houston, Texas: unexpected abundance of clonal group 307. MBio. 2017;8:e00489–17. DOIPubMedGoogle Scholar
- Giddins MJ, Macesic N, Annavajhala MK, Stump S, Khan S, McConville TH, et al. Successive emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in blaKPC-2-harboring Klebsiella pneumoniae sequence type 307 isolates. Antimicrob Agents Chemother. 2018;62:e02101–17.
- Escandón-Vargas K, Reyes S, Gutiérrez S, Villegas MV. The epidemiology of carbapenemases in Latin America and the Caribbean. Expert Rev Anti Infect Ther. 2017;15:277–97. DOIPubMedGoogle Scholar
- Pasteran FG, Otaegui L, Guerriero L, Radice G, Maggiora R, Rapoport M, et al. Klebsiella pneumoniae Carbapenemase-2, Buenos Aires, Argentina. Emerg Infect Dis. 2008;14:1178–80. DOIPubMedGoogle Scholar
- Falco A, Ramos Y, Franco E, Guzmán A, Takiff H. A cluster of KPC-2 and VIM-2-producing Klebsiella pneumoniae ST833 isolates from the pediatric service of a Venezuelan Hospital. BMC Infect Dis. 2016;16:595. DOIPubMedGoogle Scholar
- Castanheira M, Costello AJ, Deshpande LM, Jones RN. Expansion of clonal complex 258 KPC-2-producing Klebsiella pneumoniae in Latin American hospitals: report of the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother. 2012;56:1668–9, author reply 1670–1. DOIPubMedGoogle Scholar
- Robledo IE, Aquino EE, Vázquez GJ. Detection of the KPC gene in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii during a PCR-based nosocomial surveillance study in Puerto Rico. Antimicrob Agents Chemother. 2011;55:2968–70. DOIPubMedGoogle Scholar
- Morrill HJ, Pogue JM, Kaye KS, LaPlante KL. Treatment options for carbapenem-resistant Enterobacteriaceae infections. Open Forum Infect Dis. 2015;2:ofv050.
Page created: July 16, 2019
Page updated: July 16, 2019
Page reviewed: July 16, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.