Volume 25, Number 8—August 2019
Research Letter
Molecular Genotyping of Hepatitis A Virus, California, USA, 2017–2018
Figure
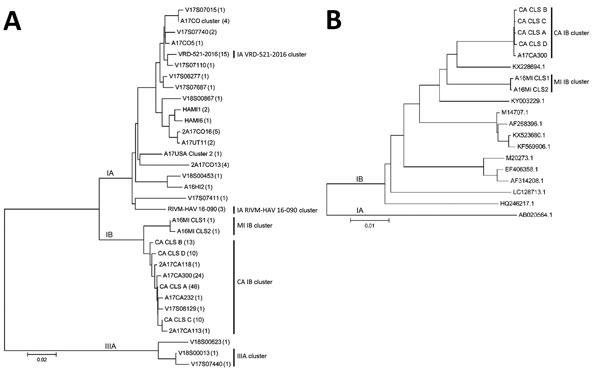
Figure. Phylogenetic analysis of HAV sequences from California, USA, and reference sequences. A) Comparison of VP1–P2B sequences obtained for 160 specimens. The number of specimens represented by each VP1–P2B sequence is indicated within parentheses. B) Comparison of nearly complete genome sequences (7,306 nt) for representative subgenotype IB cluster strains with HAV IB strain sequences found in GenBank. The genome sequence (M14707.1) represents the IB reference strain (HM175). A HAV subgenotype IA sequence (AB020564.1) was used as outlier for the analysis of the nearly complete genome sequences. Sequence alignments were performed with ClustalW (http://www.clustal.org), and the dendrograms were generated using the neighbor-joining algorithm and Kimura 2-parameter evolutionary model. Dendrogram branches corresponding to subgenotype lineage are labeled. Identified clusters of HAV are shown to the right of each dendrogram. HAV, hepatitis A virus. Scale bars indicate evolutionary distance.
1Current affiliation: Sonoma County Public Health Laboratory, Santa Rosa, California, USA.