Volume 25, Number 8—August 2019
Research Letter
Recombinant GII.Pe-GII.4 Norovirus, Thailand, 2017–2018
Figure
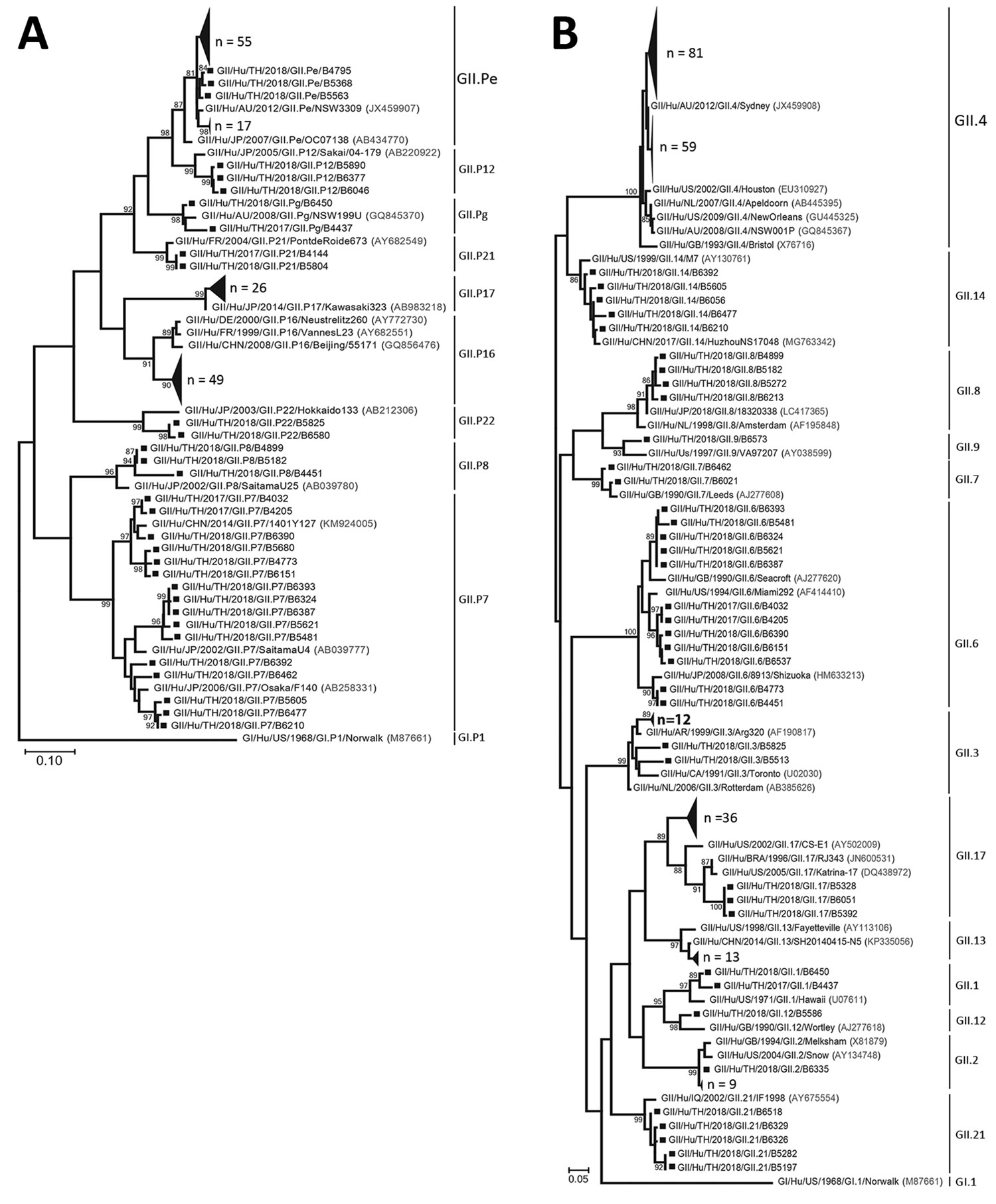
Figure. Phylogenetic trees of the norovirus GII partial-nucleotide sequences. A) Analysis of the RNA-dependent RNA polymerase (RdRp) region (380 bp). B) Analysis of the major capsid protein VP1 region (271 bp). Trees were generated by using the maximum-likelihood method with 1,000 bootstrap replicates implemented in MEGA7 (https://www.megasoftware.net). Bootstrap values >80 are indicated at the nodes. Strains of sufficient nucleotide sequence length were included in the trees (denoted individually with squares and in groups with large triangles). Reference strains are shown with accession numbers (in parentheses). Scale bars indicate nucleotide substitutions per site.
Page created: July 17, 2019
Page updated: July 17, 2019
Page reviewed: July 17, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.