Volume 25, Number 9—September 2019
Research
Whole-Genome Sequencing of Salmonella Mississippi and Typhimurium Definitive Type 160, Australia and New Zealand
Figure 5
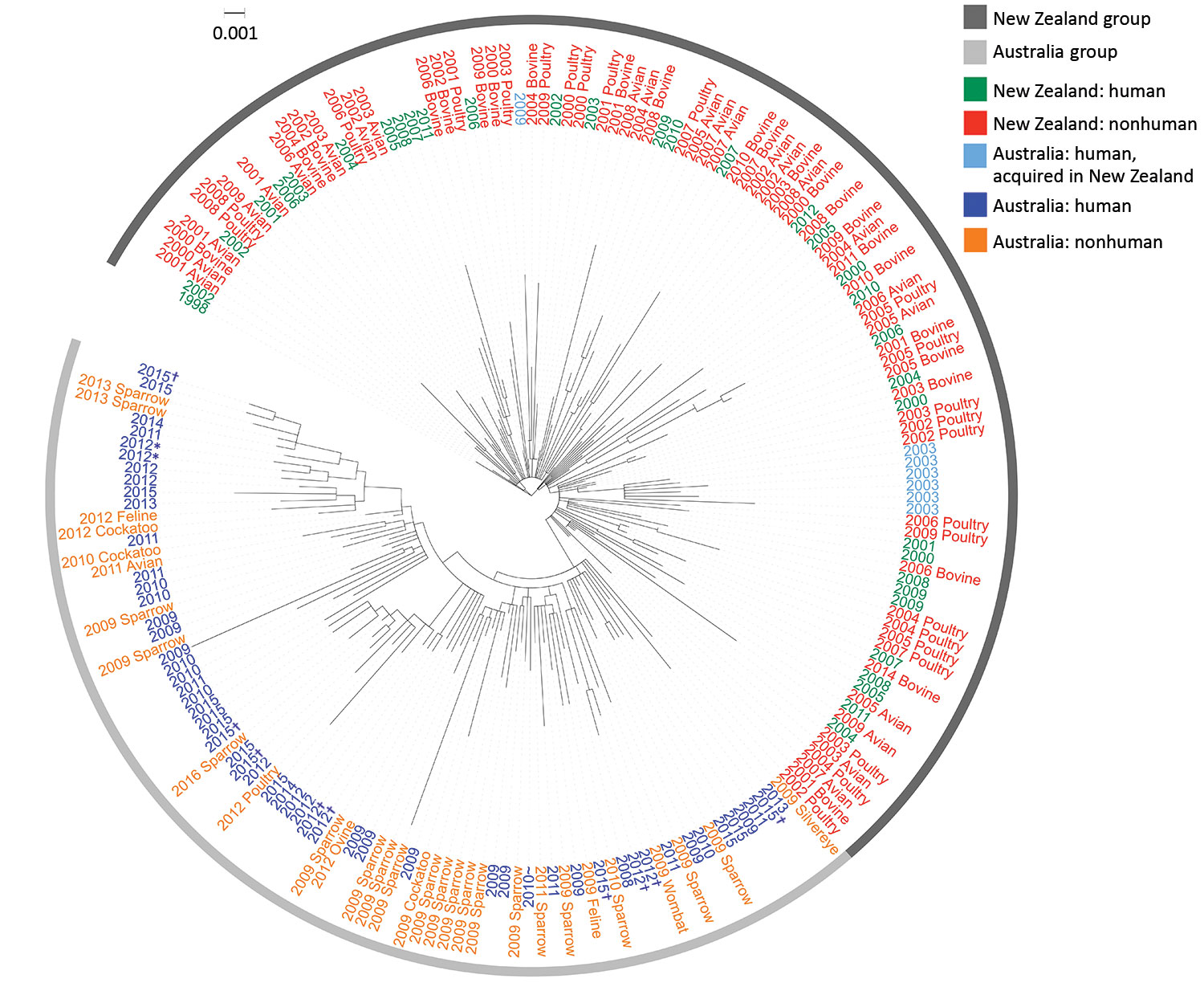
Figure 5. Maximum-likelihood phylogeny of 198 sequenced Salmonella enterica serovar Typhimurium definitive type 160 isolates from Australia and New Zealand and reference isolates, inferred from 2,203 core single-nucleotide polymorphisms, Australia and New Zealand. Nodes are labeled with isolate type and isolation year. All Australian isolates are from Tasmania unless specified otherwise. Figure created with iTOL (https://itol.embl.de). Scale bar indicates nucleotide substitutions per site. *Specimens from the same person. †Investigated as part of an epidemiologic cluster. ‡Acquired in New South Wales.
Page created: August 20, 2019
Page updated: August 20, 2019
Page reviewed: August 20, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.