Volume 25, Number 9—September 2019
Research Letter
Dengue Virus Type 1 Infection in Traveler Returning from Tanzania to Japan, 2019
Figure
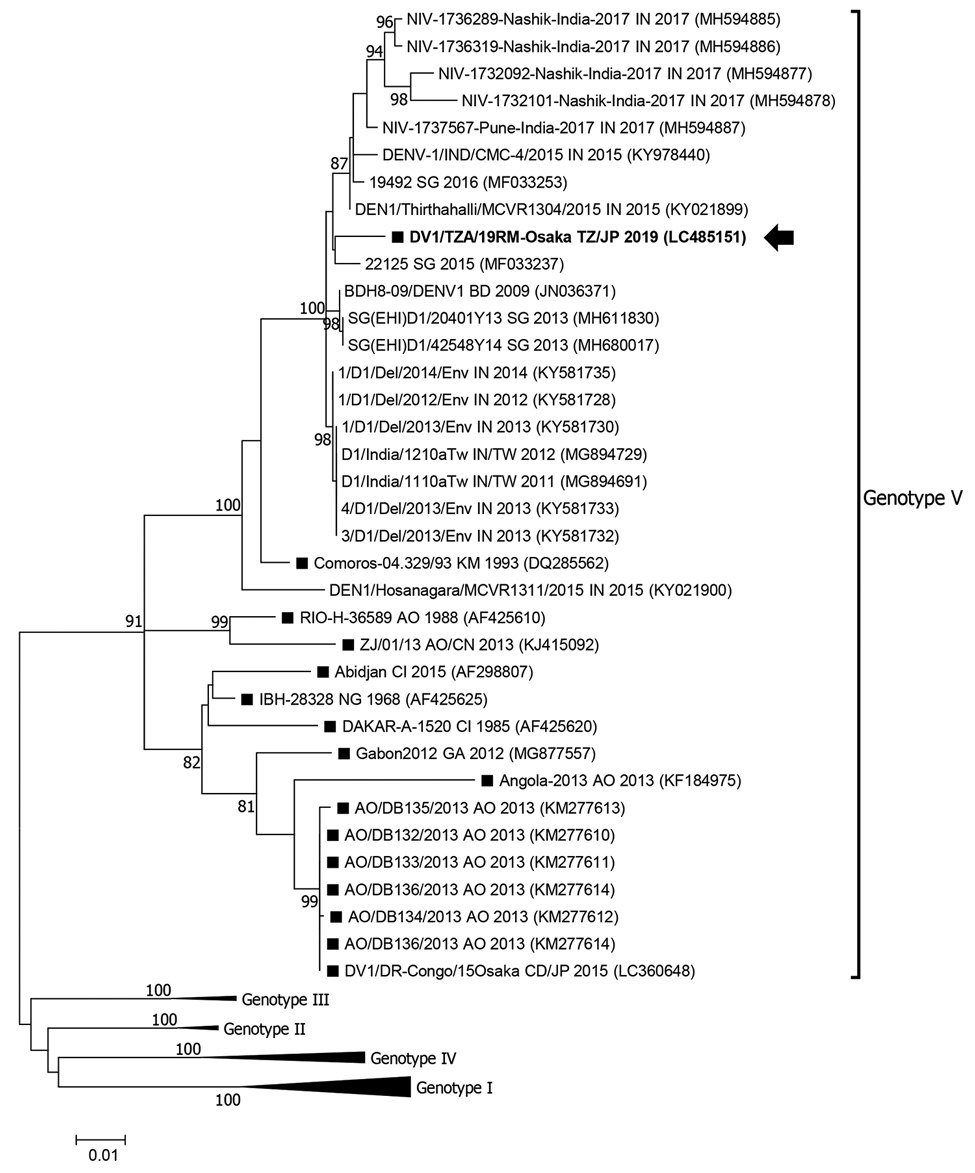
Figure. Maximum-likelihood phylogram of the envelope gene (1,485 nt) of dengue virus type 1 strains detected in Osaka, Japan, 2019 (arrow), Africa (black squares), and reference strains. Based on Bayesian information criteria, the Tamura-Nei plus gamma model was used to construct the phylogram. Numbers at the nodes indicate the bootstrap support values, which are expressed as a percentage of 1,000 replicates (values <80% are omitted). Each strain is identified by strain name, 2-letter country name abbreviation (country exported from/to, in the case of travelers), and detection year; accession numbers are shown in parentheses. Genotype I–IV branches are condensed for space. Scale bar indicates genetic distance (nucleotide substitutions per site). AO, Angola; BD, Bangladesh; CD, Democratic Republic of the Congo; CI, Côte d'Ivoire; GA, Gabon; IN, India; JP, Japan; KM, Comoros; NG, Nigeria; SG, Singapore; TW, Taiwan; TZ, Tanzania.