Volume 26, Number 1—January 2020
Dispatch
Locally Acquired Human Infection with Swine-Origin Influenza A(H3N2) Variant Virus, Australia, 2018
Figure 2
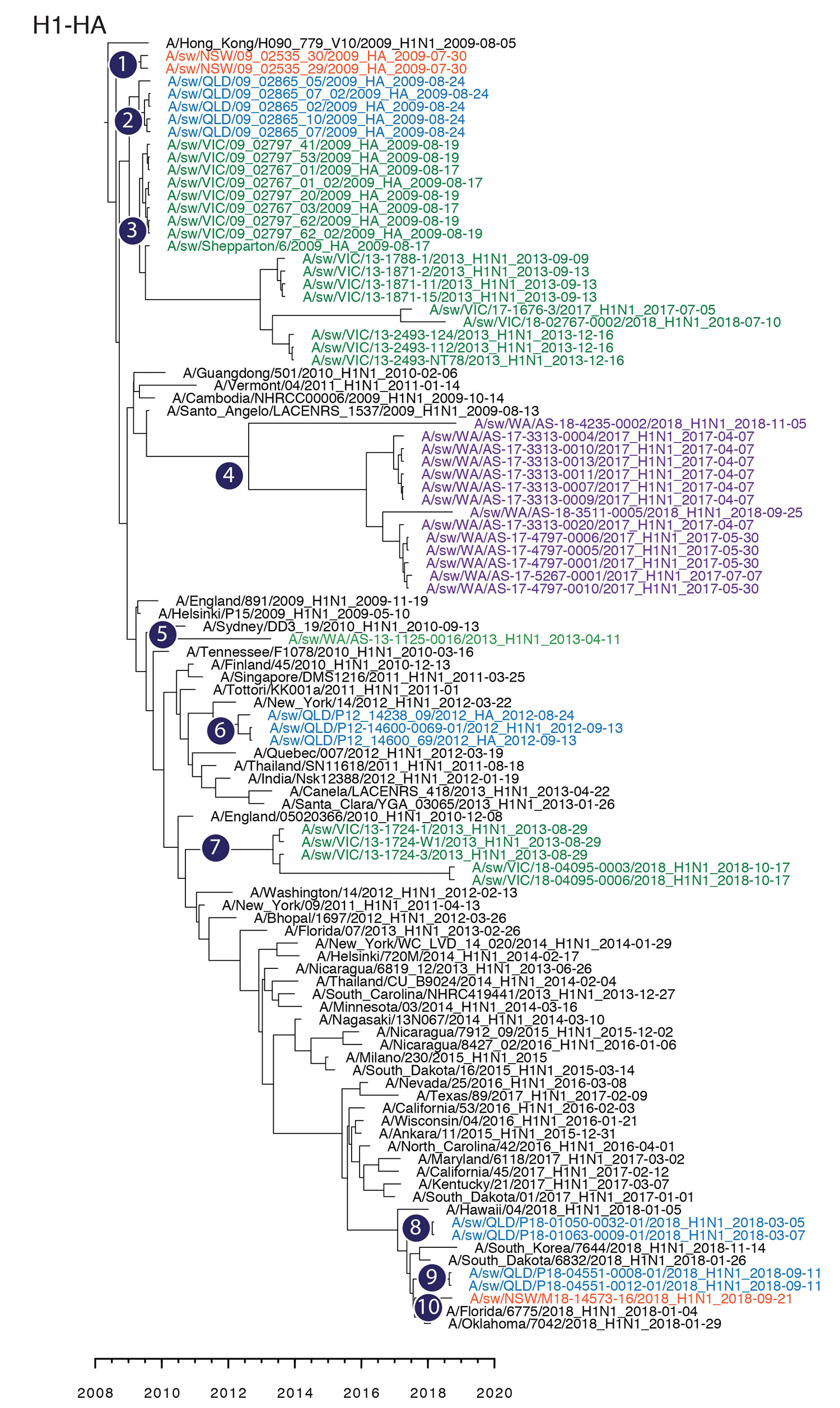
Figure 2. Maximum-clade credibility tree showing the time of emergence from humans and divergence of swine influenza A and influenza A(H1N1)pdm09 virus among swine populations in Australia. Numbers 1–10 denote inferences of individual introductions from humans to swine, and virus names are colored by state of collection: red, New South Wales; blue, Queensland; green, Victoria; purple, Western Australia. Time-scaled phylogenies were estimated by using the uncorrelated log-normal relaxed clock model (11) in a Bayesian Markov chain Monte Carlo framework in BEAST version 1.10 (https://beast.community). HA, hemagglutinin.
References
- Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–5. DOIPubMedGoogle Scholar
- Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, et al.; Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–15. DOIPubMedGoogle Scholar
- Vincent A, Awada L, Brown I, Chen H, Claes F, Dauphin G, et al. Review of influenza A virus in swine worldwide: a call for increased surveillance and research. Zoonoses Public Health. 2014;61:4–17. DOIPubMedGoogle Scholar
- Bowman AS, Nelson SW, Page SL, Nolting JM, Killian ML, Sreevatsan S, et al. Swine-to-human transmission of influenza A(H3N2) virus at agricultural fairs, Ohio, USA, 2012. Emerg Infect Dis. 2014;20:1472–80. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention (CDC). Influenza A (H3N2) variant virus-related hospitalizations: Ohio, 2012. MMWR Morb Mortal Wkly Rep. 2012;61:764–7.PubMedGoogle Scholar
- Wong FYK, Donato C, Deng YM, Teng D, Komadina N, Baas C, et al. Divergent human-origin influenza viruses detected in Australian swine populations. J Virol. 2018;92:e00316–8. DOIPubMedGoogle Scholar
- Matrosovich M, Matrosovich T, Carr J, Roberts NA, Klenk HD. Overexpression of the alpha-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J Virol. 2003;77:8418–25. DOIPubMedGoogle Scholar
- Hayden FG, Hay AJ. Emergence and transmission of influenza A viruses resistant to amantadine and rimantadine. Curr Top Microbiol Immunol. 1992;176:119–30. DOIPubMedGoogle Scholar
- Deng YM, Iannello P, Smith I, Watson J, Barr IG, Daniels P, et al. Transmission of influenza A(H1N1) 2009 pandemic viruses in Australian swine. Influenza Other Respir Viruses. 2012;6:e42–7. DOIPubMedGoogle Scholar
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. DOIPubMedGoogle Scholar
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:
e88 . DOIPubMedGoogle Scholar - Duwell MM, Blythe D, Radebaugh MW, Kough EM, Bachaus B, Crum DA, et al. Influenza A(H3N2) variant virus outbreak at three fairs —Maryland, 2017. MMWR Morb Mortal Wkly Rep. 2018;67:1169–73. DOIPubMedGoogle Scholar
- Epperson S, Jhung M, Richards S, Quinlisk P, Ball L, Moll M, et al.; Influenza A (H3N2)v Virus Investigation Team. Human infections with influenza A(H3N2) variant virus in the United States, 2011-2012. Clin Infect Dis. 2013;57(Suppl 1):S4–11. DOIPubMedGoogle Scholar
- Jhung MA, Epperson S, Biggerstaff M, Allen D, Balish A, Barnes N, et al. Outbreak of variant influenza A(H3N2) virus in the United States. Clin Infect Dis. 2013;57:1703–12. DOIPubMedGoogle Scholar
- Animal Health Australia. AUSVETPLAN manuals and documents. Influenza A viruses in swine (Version 4.0) [cited 2019 Jul 5]. https://www.animalhealthaustralia.com.au/our-publications/ausvetplan-manuals-and-documents
Page created: December 18, 2019
Page updated: December 18, 2019
Page reviewed: December 18, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.