COVID-19 Outbreak, Senegal, 2020
Ndongo Dia
1
, Ndeye Aïssatou Lakh
1, Moussa Moise Diagne, Khardiata Diallo Mbaye, Fabien Taieb, Ndeye Maguette Fall, Mamadou Alioune Barry, Daye Ka, Amary Fall, Viviane Marie Pierre Cisse Diallo, Oumar Faye, Mamadou Malado Jallow, Idrissa Dieng, Mamadou Ndiaye, Mamadou Diop, Abdoulaye Bousso, Cheikh loucoubar, Marie Khemesse Ngom Ndiaye, Christophe Peyreffite, Louise Fortes, Amadou Alpha Sall, Ousmane Faye
1, and Moussa Seydi
1
Author affiliations: Institut Pasteur, Dakar, Senegal (N. Dia, M.M. Diagne, F. Taieb, M.A. Barry, A. Fall, Oumar Faye, M.M. Jallow, I. Dieng, M. Diop, C. Ioucoubar, C. Peyreffite, A.A. Sall, Ousmane Faye, M. Seydi); Service des Maladies infectieuses de l'hôpital Fann, Dakar (N.A. Lakh, K.D. Mbaye, N.M. Fall, D. Ka, V.M.P.C. Diallo, L. Fortes); Ministère de la Santé et de l’Action Sociale (MSAS), Dakar (M. Ndiaye, A. Bousso, M.K.N. Ndiaye)
Main Article
Figure
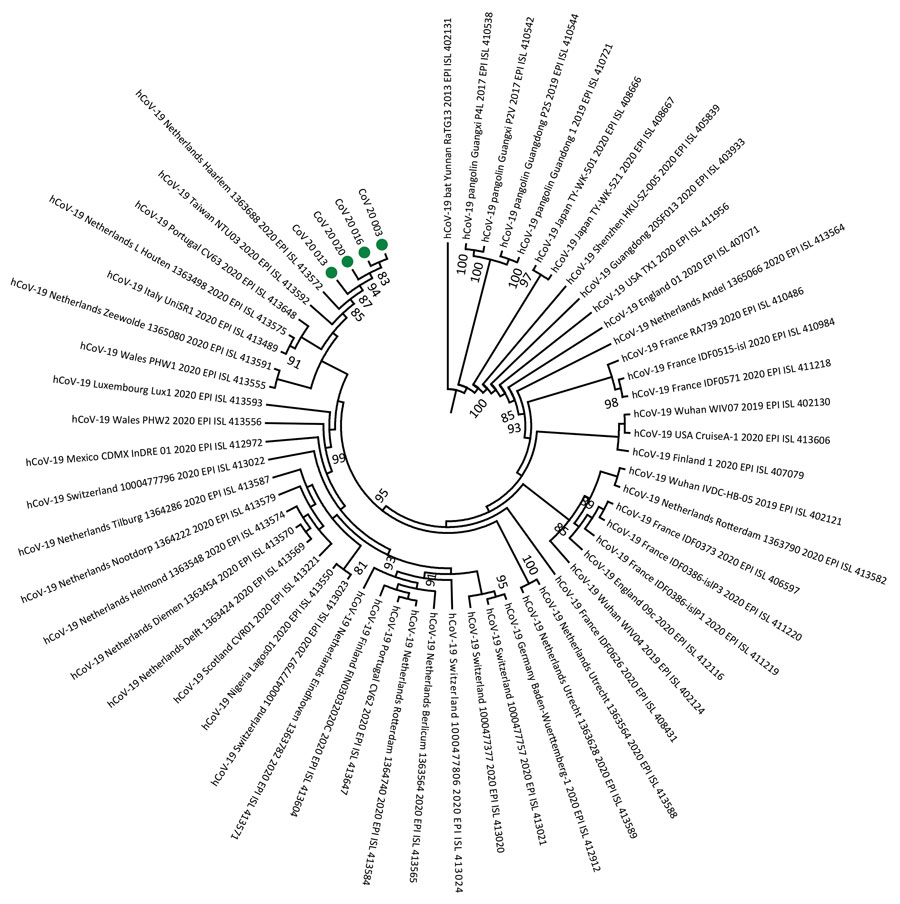
Figure. Phylogeny of 4 severe acute respiratory syndrome coronavirus 2 strains isolated from Senegal (green dots). Whole-genome nucleotide sequences were compared with 56 other genome sequences from the coronavirus disease pandemic retrieved from GenBank and GISAID (https://www.gisaid.org) databases. Sequences were aligned with MAFFT (https://mafft.cbrc.jp/alignment/server). We generated the phylogenetic tree by the maximum-likelihood method under the HKY85-gamma nucleotide substitution model using IQ-TREE (http://www.cibiv.at/software/iqtree). We assessed robustness of tree topology with 1,000 replicates; bootstrap values >75% are shown on the branches of the consensus trees. Phylogenetic analyses revealed that strains from Senegal clustered with strains from diverse origins (Europe, Asia, Latin America, and Africa). CoV, coronavirus; hCoV, human coronavirus.
Main Article
Page created: September 11, 2020
Page updated: October 19, 2020
Page reviewed: October 19, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
