Volume 26, Number 12—December 2020
Dispatch
Identification of a Novel α-herpesvirus Associated with Ulcerative Stomatitis in Donkeys
Figure 2
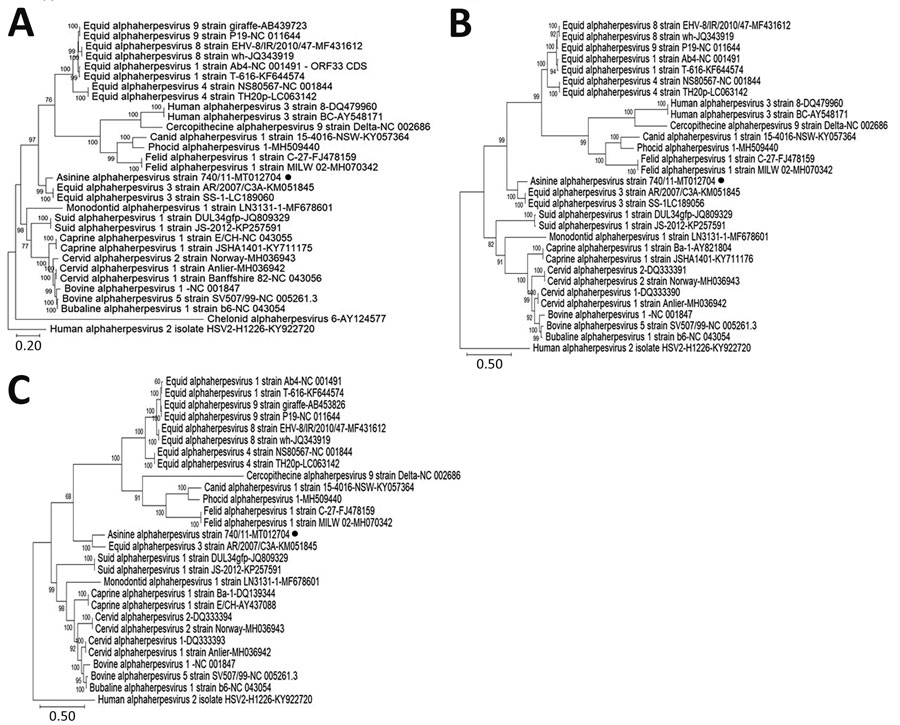
Figure 2. Phylogenetic trees based on the full-length nucleotide sequence of different target genes from the AsHV/Bari/2011/740 strain isolated from a donkey dairy herd, Bari, Italy (black dots), or retrieved from the International Committee on Taxonomy of Viruses database. A) Glycoprotein B; B) glycoprotein C; C) glycoprotein D. Posterior output of the tree was derived from maximum-likelihood inference using a general time-reversible model, a proportion of invariable sites, a gamma distribution of rate variation across sites, and a subsampling frequency of 1,000. Posterior probability values >70% are indicated on the tree nodes. Human alphaherpesvirus 2 isolate HSV-2-H1226 (genus Simplesvirus) strain (GenBank accession no. KY922720) was used as an outgroup. Scale bars indicate nucleotide substitutions per site. AsHV, asinine herpesvirus.