Volume 26, Number 12—December 2020
Dispatch
Highly Pathogenic Avian Influenza A(H7N3) Virus in Poultry, United States, 2020
Figure 2
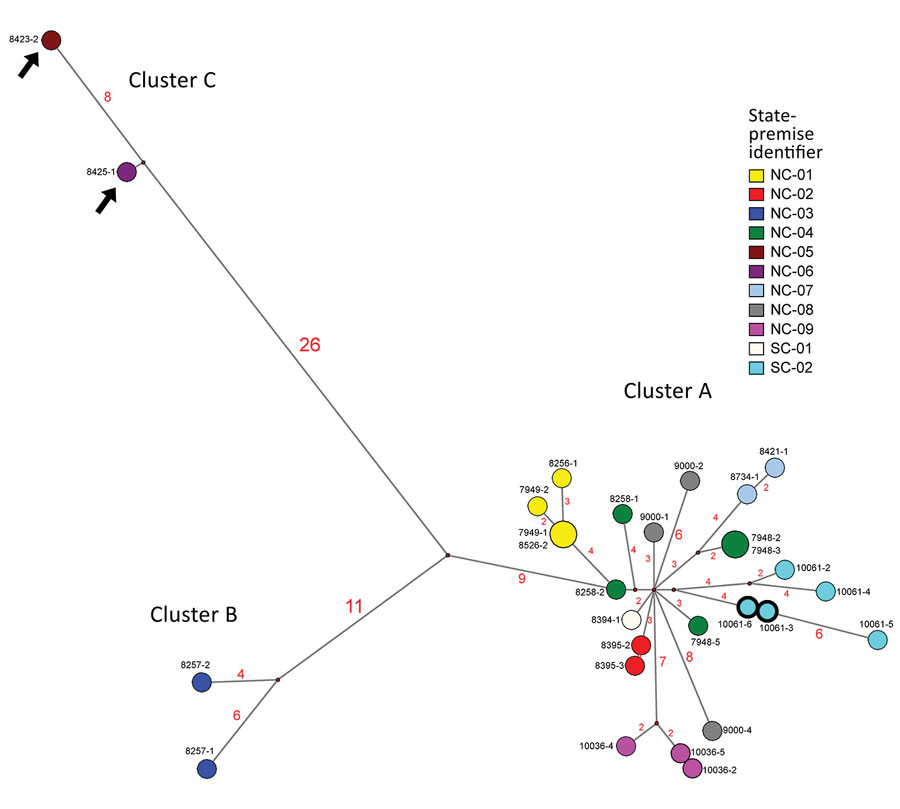
Figure 2. Median-joining phylogenetic network of the concatenated whole genome of highly pathogenic avian influenza (HPAIV) A(H7N3) viruses from South Carolina and North Carolina , USA. This network tree includes all the most parsimonious trees linking the sequences. Each unique sequence is represented by a circle sized relative to its frequency. The number of nucleotide differences between viruses is indicated on the branches. Isolates are colored according to the source premises. The black arrows indicate the H7N3 viruses with the 66-nt deletion at the neuraminidase stalk region (the NA of these 2 viruses was modified to exclude the deletion for this analysis). Both low-pathogenicity (turquoise) and HPAIV (turquoise with bolded black outline) viruses were recovered from the SC-02 premises (the hemagglutinin of the 2 HPAIVs excludes the insertion for this analysis).
1These authors contributed equally to this article.