Donor-Derived Transmission of Cryptococcus gattii sensu lato in Kidney Transplant Recipients
Daniel W.C.L. Santos, Ferry Hagen, Jacques F. Meis, Marina P. Cristelli, Laila A. Viana, Fabiola D.C. Bernardi, Hélio Tedesco-Silva, José O. Medina-Pestana, and Arnaldo L. Colombo

Author affiliations: Hospital do Rim, São Paulo, Brazil (D. W.C.L. Santos, M.P. Cristelli, L.A. Viana, H. Tedesco-Silva, J.O. Medina-Pestana); Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo (D.W.C.L. Santos, A.L. Colombo); Canisius Wilhelmina Hospital, Nijmegen, the Netherlands (F. Hagen, J.F. Meis); Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands (F. Hagen); Centre of Expertise in Mycology Radboudumc/CWZ, Nijmegen, the Netherlands (J.F. Meis); Faculdade de Ciências Médicas da Santa Casa de São Paulo, São Paulo (F.D.C. Bernardi)
Main Article
Figure
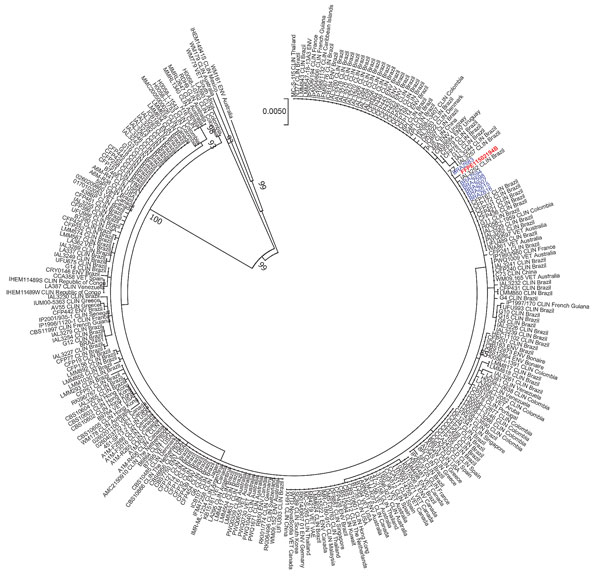
Figure. Phylogenetic maximum-likelihood analysis of 6 Cryptococcus deuterogattii isolates from 2 kidney transplant recipients in Brazil (blue), the organ donor (red), and cases from the literature (Appendix, https://wwwnc.cdc.gov/EID/article/26/6/19-1765-App1.pdf), along with reference isolates. Isolates from the 2 transplant recipients came from blood and urine; the genetic material from the donor came from a formalin-fixed paraffin-embedded brain tissue block. The GenBank accession numbers are KU642696-KU642701 (CAP59), KU642741-KU642746 (GPD1), KU642786-KU642791 (IGS1), KU642831-KU642836 (LAC1), KU642876-KU642881 (PLB1), KU642967-KU642972 (SOD1) and KU642921-KU642926 (URA5). Tree represents 1,000x bootstraps. Scale bar represents substitutions per site.
Main Article
Page created: May 19, 2020
Page updated: May 19, 2020
Page reviewed: May 19, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
