Volume 26, Number 9—September 2020
Dispatch
Chromobacterium haemolyticum Pneumonia Associated with Near-Drowning and River Water, Japan
Figure 1
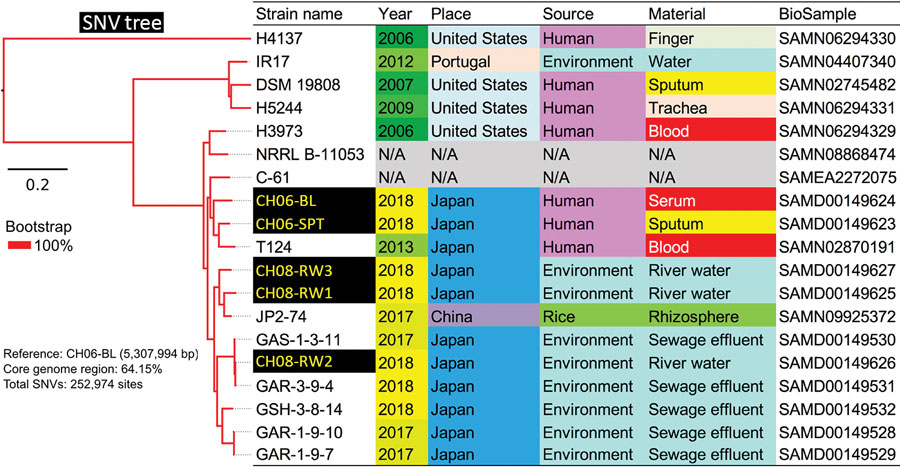
Figure 1. Core genome single-nucleotide variations in a phylogenetic analysis of 19 strains of Chromobacterium haemolyticum in a case of pneumonia associated with near-drowning in river water, Japan. In total, 252,974 SNV sites were detected in core genome region among 19 strains. The phylogenetic analysis with SNV data was constructed by maximum likelihood method. Two clinical isolates (CH06-BL and CH06-SPT) and 3 environmental isolates (CH08-RW1, CH08-RW2, and CH08-RW3) of C. haemolyticum in this study were discordant (27,867–29,491 SNVs). Scale bar indicates nucleotide substitutions per site. SNV, single nucleotide variant.
Page created: July 06, 2020
Page updated: August 19, 2020
Page reviewed: August 19, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.