Volume 26, Number 9—September 2020
Dispatch
Role of Wildlife in Emergence of Ebola Virus in Kaigbono (Likati), Democratic Republic of the Congo, 2017
Cite This Article
Citation for Media
Abstract
After the 2017 Ebola virus (EBOV) outbreak in Likati, a district in northern Democratic Republic of the Congo, we sampled small mammals from the location where the primary case-patient presumably acquired the infection. None tested positive for EBOV RNA or antibodies against EBOV, highlighting the ongoing challenge in detecting animal reservoirs for EBOV.
The animal reservoir(s) for Ebola virus (EBOV) remain unclear. Although substantial evidence suggests several bat species can host EBOV and other filoviruses (1–8), it cannot be ruled out that other, less frequently surveyed mammal groups could also host these viruses or play a role in their ecology (9). An EBOV outbreak in humans implies that EBOV had been circulating among wildlife where the primary case-patient contracted the infection. If the primary case-patient and his or her activities before becoming ill are known, this information provides an opportunity for EBOV wildlife surveillance closely focused in space and season.
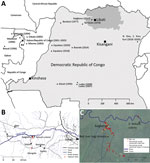
In late March 2017, signs and symptoms of hemorrhagic fever developed in an inhabitant of Kaigbono, a village in Likati district in northern Democratic Republic of the Congo (Figure 1, panel A) (10). In subsequent weeks, 2 probable and 5 confirmed cases of Ebola virus disease followed in nearby villages (Figure 1, panel B) (10,11). The World Health Organization officially declared this outbreak over on July 2, 2017. Our team arrived in Kaigbono on July 5, 2017, to investigate potential EBOV circulation among local wildlife.
The primary case-patient spent the weeks preceding his illness in and around his home village of Kaigbono, a settlement of <50 inhabitants near the Likati River (Figure 1). The village is accessible only via the Likati River or narrow forest paths. The primary case-patient often collected fish from fishermen along the river and transported it to the village.
This primary case-patient ate cooked meat from a red river hog (Potamochoerus porcus) »13 days before symptom onset. Other persons had found the dead hog in the forest ≈700 m from the village. Up to 4 Kaigbono villagers (none of whom became ill) collected the meat around the upward shoulder area of the hog, reportedly leaving the rest of the carcass because the meat touching the ground and the internal organs had already decomposed. Two of these persons prepared and cooked the meat, which subsequently was shared by ≈10–20 persons in Kaigbono, including the primary case-patient (the only person in whom febrile illness developed). On July 10 we retrieved a skull of a red river hog at the site described as the location where the abovementioned hog was found (Figure 1, panel C). Another potential zoonotic exposure occurred »7 days before symptom onset in the primary case-patient when he brought home a large bat. Other persons had hunted and killed the bat, probably at the site of a large seasonal colony of straw-colored fruit bats at the Likati River (Eidolon helvum; Figure 1, panels B, C). Villagers reported that the colony arrives annually in March and leaves in July; we observed that most of the E. helvum bats left the site during July 16–19. Given the colony’s large size (at least several thousand), most bats hunted during this season probably belong to E. helvum. However, we observed that Hypsignathus monstrosus and Epomops franqueti, other bat species in which EBOV RNA has been documented (3), also might have been occasionally hunted. The primary case-patient’s wife prepared (removed its internal organs) and grilled the bat. Only the primary case-patient ate the bat. His wife did not report a fever.
From July 6, 2017 through August 18, 2017, we trapped 476 small mammals (rodents, shrews, and bats) and acquired samples from 11 mammals hunted by local inhabitants. None of the animals showed signs of illness. Of these animals, we euthanized and collected organ samples (preserved in RNALater) of 388 (when possible, we also collected dried blood spots and oral, urogenital, and/or rectal swab specimens from these animals); collected only dried blood spots and oral, urogenital, and/or rectal swab specimens of 79; and did not sample 20. We collected bat fecal samples from plastic sheets fixed to trees underneath the E. helvum bat colony (Figure 1, panels B, C). We also swabbed the exterior of the skull of the abovementioned red river hog and extracted its molars.
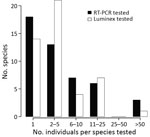
We extracted RNA from >1 organ, blood, and/or fecal samples of 419 of the 467 sampled animals and from samples swabbed from the skull and the molar pulp remains of the red river hog. We performed multiplex quantitative reverse transcription PCR (qRT-PCR) targeting EBOV and Sudan virus L gene, as previously described (12). We further tested RNA extract from 91 fecal samples and 1 urine sample collected at the E. helvum bat colony, although 47 of these samples showed signs of PCR inhibition. None of the samples of the total of 465 individual animals or environmental feces were qRT-PCR positive (Figure 2; Appendix Table).
We used a 10-antigen Luminex assay (Luminex Corporation, https://www.luminexcorp.com) to test dried blood spots of 272 animals and 92 fecal samples for antibodies against EBOV, as previously described (2,13). None of these samples could be considered positive for antibodies against EBOV (Appendix Figure).
Cytochrome b, 16S, or 12S gene PCR was attempted and Sanger sequenced on a subset (n = 334) of specimens for host species confirmation. We deposited sequences in GenBank under accession nos. MN597466–MN597893.
We distinguished 47 different mammal species (4 could not be assigned to a known species catalogued in GenBank) from 34 different genera across 359 specimens. For 268 of these, genetic information was necessary to identify the species, as species identification was not possible or not done correctly in the field (Appendix Table). Most species had low sample sizes with little power to detect low virus prevalences (Figure 2; Appendix). For an additional 67 nongenotyped animals, the genus could be unambiguously determined based on field morphology. We did not determine a genus or species for 62 animals.
Before his illness, the primary case-patient of the 2017 EBOV outbreak in Likati had eaten prepared meat from a red river hog and a fruit bat, probably E. helvum. He had contact with the uncooked carcass of the bat but not of the hog. The meat of the bat and the hog were prepared by others who had not fallen ill but whose serologic status is unknown. The hog had been dead for several days before butchering and cooking, causing us to question the infectiousness of any virus present in the meat. The susceptibility of the bat species E. helvum to EBOV is also questionable because experimental data suggests EBOV could be refractory in E. helvum cells (14). Thus, we can neither exclude nor confirm which or if either of these animals sparked the Likati 2017 EBOV outbreak.
We started collecting wildlife specimens ≈3 months after the onset of the human outbreak, a time lag potentially important to local natural transmission dynamics yet still within similar seasonal conditions. We ceased sampling 6 weeks later when seasonal changes occurred (e.g., emigration of the E. helvum bat colony). Despite this directed sampling, we did not find evidence for EBOV RNA (n = 465 animals tested) or antibodies against EBOV (n = 364) in any wildlife specimen. Because we used a qRT-PCR specific to EBOV and Sudan virus, we cannot exclude the presence of RNA of other filoviruses in the samples. However, the Luminex assay would have revealed any antibodies against related filoviruses.
As noted in previous surveillance studies following an EBOV outbreak (15; reference 1 in Appendix), the high mammal species diversity in the Congo basin rainforest, combined with the remoteness of the outbreak site, complicates the collection of a sufficient sample size for all potentially relevant taxa (Figure 2; Appendix). Furthermore, many small mammals are difficult to identify at a species or genus level on the basis of morphology alone (Appendix). Therefore, we emphasize that EBOV and other virus surveillance in small mammals requires molecular identification of the host species.
Because all human EBOV outbreaks start from a spillover event from wildlife, knowledge on exactly which wildlife species are involved in EBOV natural ecology would provide a more precise geographical and seasonal risk map for human EBOV disease outbreaks. Therefore, despite the challenges highlighted by this study, investing in increased surveillance of African forest wildlife to find EBOV reservoirs could greatly benefit public health preparedness for the devastating disease caused by this virus.
Dr. Mbala-Kingebeni is a researcher at the Institut National de Recherche Biomédicale and University of Kinshasa, Democratic Republic of Congo. His research interests include characterization of hosts that harbor zoonotic pathogens.
Acknowledgments
We thank Lieselotte Cnops for logistic support and 3 anonymous reviewers for useful comments on the manuscript.
This work was supported by funds from the VLIR-UOS University Development Cooperation (Institutional University Cooperation); University of Antwerp; the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health; Institut National de la Santé et de la Recherche Médicale/Ebola Task Force; REACTing (REsearch and ACTion targeting emerging infectious diseases); EBO-SURSY project funded by European Union; Christophe Mérieux Prize 2015 awarded to J.J.M.T.; Institut de Recherche pour le Développement; German Research Council (grant no. LE1813/7-1); the Global Health Protection Programme supported by the Federal Ministry of Health on the basis of a decision by the German Bundestag; and the Department of Economy, Science and Innovation EWI from the Flemish Government (Belgium). S.G. was supported by an OUTGOING [PEGASUS]2 Marie Skłodowska-Curie fellowship of the Research Foundation–Flanders (grant no. 12T1117N). E.V. was financially supported by the Operational Directorate of Taxonomy and Phylogeny and Capacities for Biodiversity and Sustainable Development at the Royal Belgian Institute of Natural Sciences. K.K.A. was supported by a grant from Flanders Innovation & Entrepreneurship.
References
- Pourrut X, Souris M, Towner JS, Rollin PE, Nichol ST, Gonzalez JP, et al. Large serological survey showing cocirculation of Ebola and Marburg viruses in Gabonese bat populations, and a high seroprevalence of both viruses in Rousettus aegyptiacus. BMC Infect Dis. 2009;9:159. DOIPubMedGoogle Scholar
- De Nys HM, Kingebeni PM, Keita AK, Butel C, Thaurignac G, Villabona-Arenas CJ, et al. Survey of Ebola viruses in frugivorous and insectivorous bats in Guinea, Cameroon, and the Democratic Republic of the Congo, 2015–2017. Emerg Infect Dis. 2018;24:2228–40. DOIPubMedGoogle Scholar
- Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, et al. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–6. DOIPubMedGoogle Scholar
- Towner JS, Amman BR, Sealy TK, Carroll SA, Comer JA, Kemp A, et al. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 2009;5:
e1000536 . DOIPubMedGoogle Scholar - Negredo A, Palacios G, Vázquez-Morón S, González F, Dopazo H, Molero F, et al. Discovery of an ebolavirus-like filovirus in europe. PLoS Pathog. 2011;7:
e1002304 . DOIPubMedGoogle Scholar - He B, Feng Y, Zhang H, Xu L, Yang W, Zhang Y, et al. Filovirus RNA in fruit bats, China. Emerg Infect Dis. 2015;21:1675–7. DOIPubMedGoogle Scholar
- Goldstein T, Anthony SJ, Gbakima A, Bird BH, Bangura J, Tremeau-Bravard A, et al. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat Microbiol. 2018;3:1084–9. DOIPubMedGoogle Scholar
- Yang XL, Tan CW, Anderson DE, Jiang RD, Li B, Zhang W, et al. Characterization of a filovirus (Měnglà virus) from Rousettus bats in China. Nat Microbiol. 2019;4:390–5. DOIPubMedGoogle Scholar
- Leendertz SA, Gogarten JF, Düx A, Calvignac-Spencer S, Leendertz FH. Assessing the evidence supporting fruit bats as the primary reservoirs for Ebola viruses. EcoHealth. 2016;13:18–25. DOIPubMedGoogle Scholar
- Nsio J, Kapetshi J, Makiala S, Raymond F, Tshapenda G, Boucher N, et al. Outbreak of Ebola virus disease in northern Democratic Republic of Congo. J Infect Dis. 2017;2019:3 https://academic.oup.com/jid/article/221/5/701/5426903.PubMedGoogle Scholar
- World Health Organization. Ebola Virus Disease Democratic Republic of Congo. External situation report 10. 2017 May 23. [cited 2017 May 23]. https://apps.who.int/iris/bitstream/handle/10665/255565/EbolaDRC-24052017.pdf.
- de Wit E, Rosenke K, Fischer RJ, Marzi A, Prescott J, Bushmaker T, et al. Ebola laboratory response at the Eternal Love Winning Africa campus, Monrovia, Liberia, 2014–2015. J Infect Dis. 2016;214(suppl 3):S169–76. DOIPubMedGoogle Scholar
- Ayouba A, Touré A, Butel C, Keita AK, Binetruy F, Sow MS, et al. Development of a sensitive and specific serological assay based on Luminex technology for detection of antibodies to Zaire Ebola virus. J Clin Microbiol. 2016;55:165–76. DOIPubMedGoogle Scholar
- Ng M, Ndungo E, Kaczmarek ME, Herbert AS, Binger T, Kuehne AI, et al. Filovirus receptor NPC1 contributes to species-specific patterns of ebolavirus susceptibility in bats. eLife. 2015;4:4. DOIPubMedGoogle Scholar
- Leirs H, Mills JN, Krebs JW, Childs JE, Akaibe D, Woollen N, et al. Search for the Ebola virus reservoir in Kikwit, Democratic Republic of the Congo: reflections on a vertebrate collection. J Infect Dis. 1999;179(Suppl 1):S155–63. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: July 29, 2020
1These authors contributed equally to this article.
Table of Contents – Volume 26, Number 9—September 2020
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Sophie Gryseels, Clinical and Epidemiological Virology, Rega Institute, KU Leuven, Herestraat 49, 3000 Leuven, Belgium
Top