Volume 26, Number 9—September 2020
Research Letter
Effect of Environmental Conditions on SARS-CoV-2 Stability in Human Nasal Mucus and Sputum
Figure
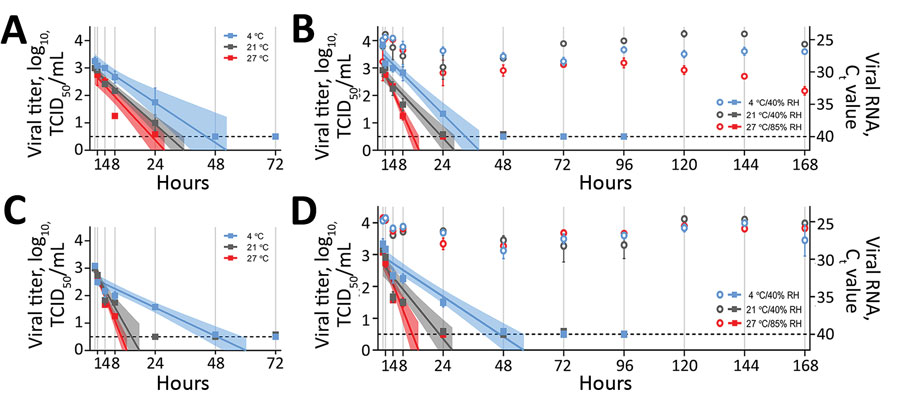
Figure. Stability of severe acute respiratory syndrome coronavirus 2 over time in human nasal mucus and sputum under different environmental conditions: liquid nasal mucus (A), surface nasal mucus (B), liquid sputum (C), and surface sputum (D). For panels B and D, the squares correspond to viral titer on the left y-axis, and the circles correspond to viral RNA (Ct value) on the right y-axis. We collected samples in 1 mL media for each condition at 0, 1, 4, 8, and 24 hours, then daily for 7 days and performed end-point titrations in quadruplicate on Vero E6 cells and made calculations using the Spearman-Kärber method. We log10-transformed and fit titers with linear regression models, including 95% CIs (shaded area around lines of best fit), by using GraphPad Prism 8 (https://www.graphpad.com). We extracted aliquots of collected surface samples by using the QIAamp Viral RNA Mini Kit (QIAGEN, https://www.qiagen.com) and analyzed them for the presence of viral RNA by using quantitative reverse transcription PCR targeting the E gene. For both viral titers and Ct values, plots show means of 3 replicates with SE. The limit of detection for each experimental condition was 100.5 TCID50/mL for viral titer and 40 for Ct value and is indicated by the dashed line. Relative humidity is not applicable to liquid samples (panels A and C), which were in sealed tubes. Ct, cycle threshold; RH, relative humidity; TCID50/mL, 50% tissue culture infective dose/mL.