Volume 27, Number 1—January 2021
Dispatch
Emerging Human Metapneumovirus Gene Duplication Variants in Patients with Severe Acute Respiratory Infection, China, 2017–2019
Cite This Article
Citation for Media
Abstract
We detected human metapneumovirus (HMPV) in 72 (7.1%) of 1,021 patients hospitalized with severe acute respiratory infection in Luohe, China, during 2017–2019. We detected HMPV most frequently in young children and less often in adults. HMPV genotype A2c variants 111 nt and 180 nt duplications predominated, demonstrating their continuing geographic spread.
Human metapneumovirus (HMPV; family Pneumoviridae, genus Metapneumovirus) is a major cause of acute respiratory tract infections, especially in children and elderly persons (1,2). Its genome is ≈13.2 kb, containing 8 genes encoding 9 proteins. The G gene, around 654–867 nt acids sequence length, is the most variable nucleotide sequence in the whole genome of HMPV and has been widely used to study HMPV genetic variation (3–5). HMPV has 1 serotype with 2 subgroups A and B, further divided into 5 genotypes, including A1, A2a, A2b, B1, and B2, based mainly on variations in the G gene (4,6,7). Recently, unique HMPV variants possessing a 180 nt duplication (nt-dup) in the G gene, first reported in Spain, and a 111 nt-dup in the G gene, first reported Japan (5,8,9), followed by Croatia and Guangdong, China (10,11). These variants were clustered in the A2c lineage of the phylogenetic tree (11). In this study, we investigated the prevalence of HMPV associated with patients with severe acute respiratory infection (SARI) and identified genetic variations in the G gene of HMPV in Luohe, in Henan Province, China, during 2017–2019.
Luohe is a city of 2.8 million people in Henan Province, northern China. In this study, we collected throat swab specimens and clinical data from 1,021 patients with SARI admitted to Luohe Central Hospital during October 2017–June 2019. Patients were 1 month–95 (median 3) years of age; most (76.7%) of the SARI cases involved children <5 years old. All swab specimens were tested by multiplex real-time reverse transcription PCR assay using a nucleic acid diagnostic kit (Kinghawk, http://www.kinghawk828.com), which identifies HMPV, influenza types A and B, human respiratory syncytial virus (RSV), human coronaviruses, human rhinovirus and enterovirus, human adenovirus, human parainfluenza viruses, and human bocavirus.
Overall, 83.2% (849/1021) of SARI patients were positive for >1 respiratory viruses; human adenovirus was the predominant virus identified, at 22% (225/1021). HMPV was identified in 7.1% (72/1021) of SARI patients, consistent with other studies that identified HMPV in 6%–12% of SARI cases (12,13). The proportion of HMPV-positive patients with co-detected respiratory viruses was 63.9% (46/72). The rates of HMPV positivity decreased gradually with age, from 9.1% in those <2 years old to 0.9% in adults (Table 1). The epidemic season for HMPV lasted from November through May or June, with the peak number of cases occurring in March and May in 2018 and March in 2019 (Figure 1). Most (98.5%) HMPV-positive patients in this study exhibited cough or dyspnea and were diagnosed with bronchopneumonia (74.6%). No clear differences in clinical signs and symptoms were apparent among the patients infected with duplication variants compared with other HMPV viruses.
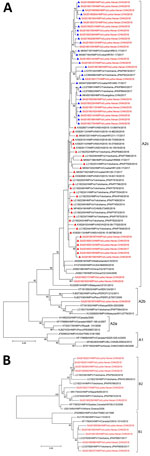
Forty-three entire coding-region sequences of the G gene were obtained from 72 HMPV-positive samples, as described elsewhere (3,5,6) (Table 2). The nucleotide sequences generated in this study were submitted to GenBank (accession nos. MN944056–97). We selected 86 subgroup A sequences (54 from GenBank and 32 from this study) and 23 subgroup B sequences (12 from GenBank and 11 from this study) to construct the phylogenetic tree by maximum-likelihood method (Figure 2). The HMPV sequences obtained in this study were clustered into 4 genotypes, including A2c, A2b, B1, and B2 (Table 2; Figure 2). Twenty-eight out of 31 A2c sequences were grouped together in 1 distinct cluster with the duplication variants, mainly detected in Spain, Japan, Croatia, and Guangzhou, China. In the sequence alignment comparison analysis, we identified 22 viruses containing 111 nt-dup variants and 6 viruses contained 180 nt-dup variants in the G gene. The 111 nt-dup variants were detected in Luohe in 2018 and continued to spread in 2019, while 180 nt-dup variants were detected during 2017 and 2018 (Table 2). The 111 nt-dup variants were separated from the 180 nt-dup variants in the phylogenetic tree, which indicates that the 2 duplication variants evolved by independent patterns in different evolutionary lineages (Figure 2, panel A).
The twenty-two 111 nt-dup variants circulating in Luohe during 2018–2019 were closely related to the duplication-variant viruses identified in Japan, Croatia, and Guangdong, China, in 2017, with nucleotide identity of 95.7% to 100%. Six 180 nt-dup variants circulating in Luohe during 2017–2018 were clustered in 1 small cluster with high nucleotide identity at 99.1% and 100% and closely related to the duplication viruses identified in Japan, Croatia, Spain, and Beijing, China, during 2014–2017 (Figure 2, panel A). This finding indicates that the 111 nt-dup and 180 nt-dup variants have already spread and might spread extensively throughout the world.
Our findings show that 7.1% of SARI patients in the Luohe study tested positive for HMPV during 2017–2019. The rate of HMPV-positivity decreased gradually with patient age. Among the HMPV patients, 63.9% were co-infected with other respiratory viruses. Four different genotypes were cocirculating, including A2b, A2c, B1, and B2. We identified most of the A2c viruses as duplication variants, including the 111 nt-dup and 180 nt-dup variants, and found that these 2 variants were the predominant viruses circulating in Luohe during 2017–2019.
The 111 nt-dup and 180 nt-dup variants have also been detected in different regions of the world, including in Spain, Japan, Croatia, and Guangzhou, China, in recent years (5,8–11). Similar duplications in the G gene have been reported in HRSV, another member of the family Pneumoviridae. The HRSV duplication variants of both BA9 and ON1 have rapidly spread globally, becoming the predominant viruses in many countries for many years (14,15). These findings suggest that the emerging HMPV 111 nt-dup and 180 nt-dup variants might also become predominant viruses throughout the world.
There were some limitations in this study. The epidemic seasons were not covered for the entire 3 years, and only 1 hospital was involved in this study. Continuous surveillance will be required to determine whether these novel HMPV 111 nt-dup and 180 nt-dup variants will persist as predominant viruses and have a wider geographic distribution in the future.
Although no difference in clinical symptoms was observed between the patients infected with HMPV duplication variants and those with non–duplication-variant viruses in this study, the increased transmission frequency of the duplication variants suggests a role for duplication in the G gene in potentially expanding its transmission. Therefore, further study is needed to clarify if and how the duplications result in an evolutionary advantage for HMPV.
Dr. Xie is a master degree student at the China CDC. Her research focuses on the epidemiology and genetic variations of human metapneumovirus and human respiratory syncytial virus.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (project no. 81772195), and the Key Technologies R&D Program of the National Ministry of Science (2018ZX10713002, 2018ZX10711001, 2017ZX10104001-002, and 2018ZX10713001-003).
References
- Esposito S, Mastrolia MV. Metapneumovirus infections and respiratory complications. Semin Respir Crit Care Med. 2016;37:512–21. DOIPubMedGoogle Scholar
- Rima B, Collins P, Easton A, Fouchier R, Kurath G, Lamb RA, et al.; Ictv Report Consortium. ICTV virus taxonomy profile: Pneumoviridae. J Gen Virol. 2017;98:2912–3. DOIPubMedGoogle Scholar
- van den Hoogen BG, Herfst S, Sprong L, Cane PA, Forleo-Neto E, de Swart RL, et al. Antigenic and genetic variability of human metapneumoviruses. Emerg Infect Dis. 2004;10:658–66. DOIPubMedGoogle Scholar
- Knipe DM, Howley PM, editors. Fields Virology. 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2013.
- Saikusa M, Kawakami C, Nao N, Takeda M, Usuku S, Sasao T, et al. 180-nucleotide duplication in the G gene of Human metapneumovirus A2b subgroup strains circulating in Yokohama City, Japan, since 2014. Front Microbiol. 2017;8:402. DOIPubMedGoogle Scholar
- Huck B, Scharf G, Neumann-Haefelin D, Puppe W, Weigl J, Falcone V. Novel human metapneumovirus sublineage. Emerg Infect Dis. 2006;12:147–50. DOIPubMedGoogle Scholar
- Kim HR, Cho AR, Lee MK, Yun SW, Kim TH. Genotype variability and clinical features of human metapneumovirus isolated from Korean children, 2007 to 2010. J Mol Diagn. 2012;14:61–4. DOIPubMedGoogle Scholar
- Piñana M, Vila J, Gimferrer L, Valls M, Andrés C, Codina MG, et al. Novel human metapneumovirus with a 180-nucleotide duplication in the G gene. Future Microbiol. 2017;12:565–71. DOIPubMedGoogle Scholar
- Saikusa M, Nao N, Kawakami C, Usuku S, Tanaka N, Tahara M, et al. Predominant detection of the subgroup A2b human metapneumovirus strain with a 111-nucleotide duplication in the G gene in Yokohama City, Japan in 2018. Jpn J Infect Dis. 2019;72:350–2. DOIPubMedGoogle Scholar
- Jagusic M, Slovic A, Ivancic-Jelecki J, Ljubin-Sternak S, Vilibić-Čavlek T, Tabain I, et al. Molecular epidemiology of human respiratory syncytial virus and human metapneumovirus in hospitalized children with acute respiratory infections in Croatia, 2014-2017. Infect Genet Evol. 2019;76:
104039 . DOIPubMedGoogle Scholar - Yi L, Zou L, Peng J, Yu J, Song Y, Liang L, et al. Epidemiology, evolution and transmission of human metapneumovirus in Guangzhou China, 2013-2017. Sci Rep. 2019;9:14022. DOIPubMedGoogle Scholar
- Al Amad MA, Al Mahaqri AA, Al Serouri AA, Khader YS. Severe acute respiratory infections with influenza and noninfluenza respiratory viruses: Yemen, 2011–2016. Inquiry. 2019;56:
46958019850731 . DOIGoogle Scholar - Breiman RF, Cosmas L, Njenga M, Williamson J, Mott JA, Katz MA, et al. Severe acute respiratory infection in children in a densely populated urban slum in Kenya, 2007-2011. BMC Infect Dis. 2015;15:95. DOIPubMedGoogle Scholar
- Song J, Wang H, Shi J, Cui A, Huang Y, Sun L, et al. Emergence of BA9 genotype of human respiratory syncytial virus subgroup B in China from 2006 to 2014. Sci Rep. 2017;7:16765. DOIPubMedGoogle Scholar
- Song J, Zhang Y, Wang H, Shi J, Sun L, Zhang X, et al. Emergence of ON1 genotype of human respiratory syncytial virus subgroup A in China between 2011 and 2015. Sci Rep. 2017;7:5501. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: December 17, 2020
Table of Contents – Volume 27, Number 1—January 2021
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Yan Zhang and Wenbo Xu, National Institute for Viral Disease Control and Prevention, China Center for Disease Control and Prevention, No. 155, Changbai Road, Changping District, Beijing 102206, China
Top