Volume 27, Number 1—January 2021
Dispatch
Emerging Human Metapneumovirus Gene Duplication Variants in Patients with Severe Acute Respiratory Infection, China, 2017–2019
Figure 2
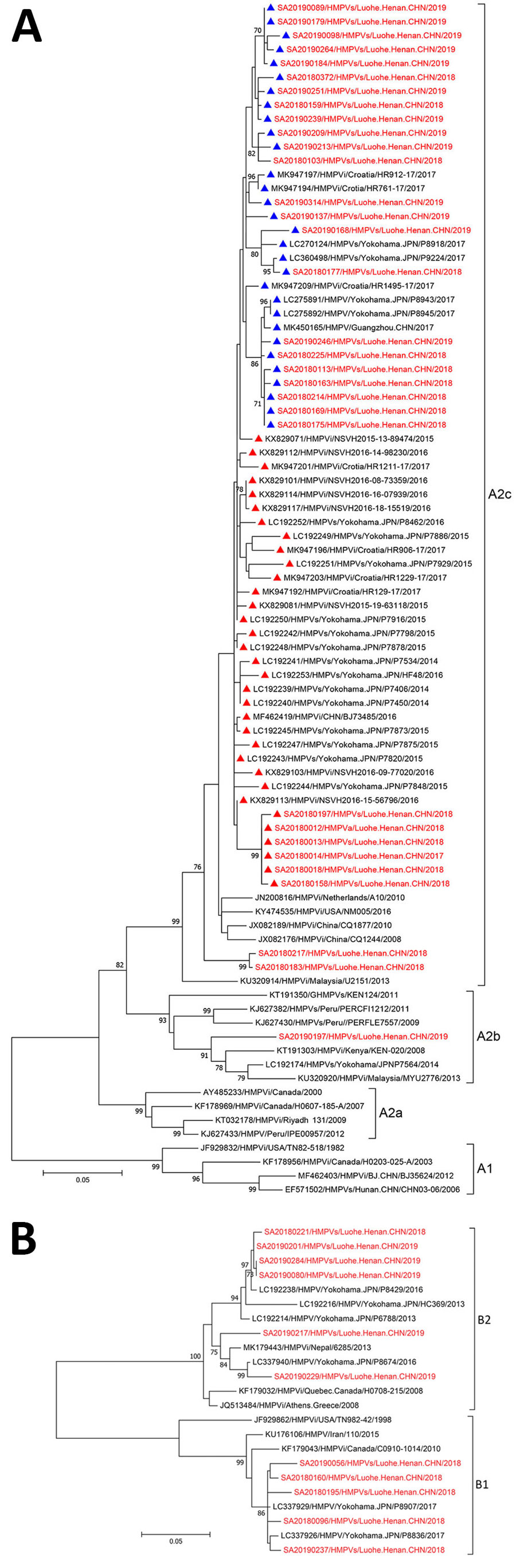
Figure 2. Phylogenetic tree generated by maximum-likelihood method of HMPV G gene sequences (red) from patients with SARI admitted to Luohe Central Hospital, Luohe, China, during October 2017–June 2019, and reference sequences (black). A) Phylogenetic tree for the representative viruses of subgroup A (A1, A2a, A2b and A2c), including 54 representative G gene sequences of HMPV from GenBank and 32 from this study. The strains with 111 nt duplication are indicated by blue triangles; strains with 180 nt duplication are indicated by red triangles. 0.05 scale bar represents 5% genetic distances for G genes (0.05 substitutions per site). B) Phylogenetic tree for the representative viruses of the B1 and B2 genotypes, including 12 representative entire coding regions of HPMV G gene sequences from GenBank and 11 from this study. The numbers at the branch nodes indicate the statistical significance, calculated with 1,000-replication bootstrapping. The nomenclature of HMPV in this study includes the following information: the source of sequences (“s” after HMPV indicates a specimen source of sequences; “i” indicates isolates), location where the viruses were detected (city, province, and country), and collection year. The GenBank accession number was added before the nomenclature of each sequence. 0.05 scale bar represents 5% genetic distances for G genes (0.05 substitutions per site). HMPV, human metapneumovirus.