Volume 27, Number 10—October 2021
Dispatch
Genetic Diversity of SARS-CoV-2 among Travelers Arriving in Hong Kong
Figure
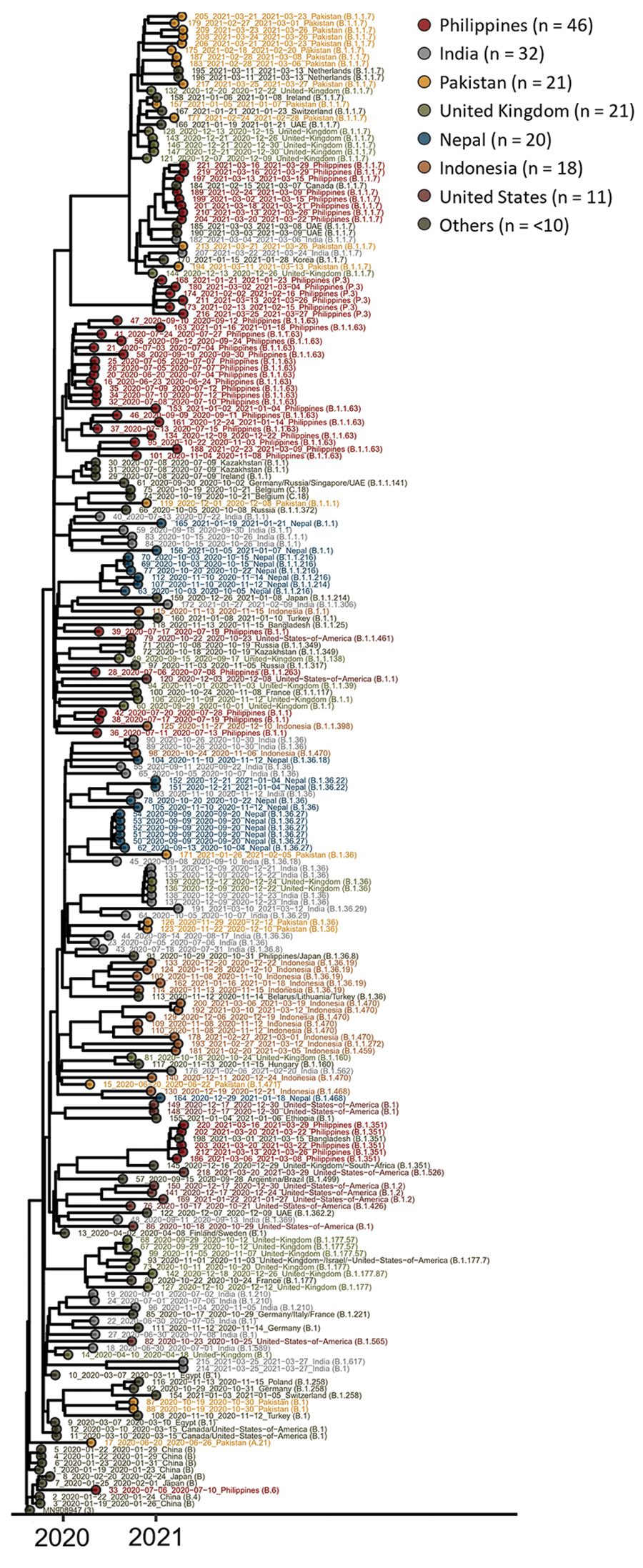
Figure. Maximum clade credibility phylogenetic tree of sequences of severe acute respiratory syndrome coronavirus 2 imported to Hong Kong, January 2020–March 2021 (Appendix 1). Date, country, and lineage information are provided.
1These first authors contributed equally to this article.
Page created: June 29, 2021
Page updated: September 19, 2021
Page reviewed: September 19, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.