Volume 27, Number 10—October 2021
Research Letter
Human Infection with Avian Influenza A(H9N2) Virus, Cambodia, February 2021
Cite This Article
Citation for Media
Abstract
In February 2021, routine sentinel surveillance for influenza-like illness in Cambodia detected a human avian influenza A(H9N2) virus infection. Investigations identified no recent H9N2 virus infections in 43 close contacts. One chicken sample from the infected child’s house was positive for H9N2 virus and genetically similar to the human virus.
Low pathogenicity avian influenza virus subtype A(H9N2) is endemic in poultry in Asia, the Middle East, and Africa (1). These viruses do not cause mass mortality in poultry but can cause substantial negative economic impacts (2). H9N2 viruses also have zoonotic potential; 74 human infections were reported from 1998 through early 2021 (1,3,4), mainly in children with a history of poultry exposure. The internal gene cassettes of H9N2 viruses contribute to human adaptation of avian influenza viruses (AIV) strains, as exemplified by the fifth wave of the H7N9 epidemic in China (5). Because of broad host range, global distribution, and reassortment capability, risk assessments indicate existing H9N2 viruses as a moderate pandemic risk (2,6).
H9N2 viruses circulate endemically in poultry from Cambodia, as they do in Bangladesh, Vietnam, and China (7,8). Seventy-five percent of AIVs detected in chickens in Cambodia in live bird markets (LBMs) are H9N2 (7). Whereas serosurveys indicate persons in Cambodia with poultry contact are exposed to H9N2, active human infections have not been detected (9,10).
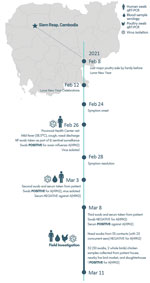
On February 26, 2021, a 3-year-old boy living in Prasat Bakong, near Siem Reap, Cambodia, was taken to an outpatient clinic for influenza-like illness with onset of symptoms on February 24 (Figure). A nasopharyngeal sample obtained as part of influenza-like illness sentinel surveillance tested positive for influenza A at the National Institute of Public Health (Phnom Penh, Cambodia) but was unsubtypable for human seasonal, H5, or H7 subtypes (Appendix 1 reference 11). The sample tested positive for H9N2 at the National Influenza Centre, Institute Pasteur du Cambodge (Phnom Penh) (7); a second specimen obtained on March 3 confirmed H9N2 infection. Viruses in the 2 samples were successfully isolated in embryonated chicken eggs and confirmed as H9N2; the second isolate was named A/Cambodia/21020301/2021. Hemagglutination inhibition testing confirmed infection by seroconversion. The serum sample taken on March 3 (7 days postonset) tested negative and then tested positive on March 10 (14 days postonset; hemagglutination inhibition = 240) (Appendix 1 reference 12).
A joint One Health investigation was undertaken during March 8–11, 2021, by the Cambodia Centers for Disease Control and Ministry of Health, National Animal Health and Production Research Institute, and provincial divisions with support from the United States Centers for Disease Control and Prevention, the World Health Organization, Food and Agriculture Organization, and Institute Pasteur du Cambodge. The infected child lives in a mainly agrarian village ≈20 km from Siem Reap encompassing 121 households and 502 inhabitants; 80% of families conduct small-scale backyard poultry farming. The child’s residence includes a chicken enclosure containing ≈50 chickens surrounded by nylon net. The child played within the enclosure and accompanied adults during feeding time. Poultry production, trading, and AIV prevalence in LBMs increase during national festival periods (7); a major sale took place on February 8, 2021, before Lunar New Year.
Forty-three close contacts were identified in 9 households (21 persons <15 and 22 persons ≥15 years of age; 53% female). Nine persons from 5 households reported respiratory symptoms immediately before or during the investigation. Nasal swab samples from 33/43 contacts (including all symptomatic persons), and concurrent serum samples from 23/43 contacts (including 8/9 symptomatic persons) showed neither H9N2 infection nor seroconversion. Health education on safe poultry farming practices was provided to representatives of close contact families.
Investigations of poultry in the index house, village, local LBMs, and slaughterhouses yielded 50 tracheal/cloacal samples and 2 whole birds. One H9N2 virus was detected from a chicken at the infected child’s house, isolated in embryonated chicken eggs, and subsequently named A/chicken/Cambodia/f0318251/2021. No poultry die-offs were reported in February 2021.
We sequenced the hemagglutinin and neuraminidase genes with Oxford Nanopore Technologies (Nanopore, https://nanoporetech.com) (Appendix 1 Figure). The hemagglutinin genes of A/Cambodia/21020301/2021 and A/chicken/Cambodia/f0318251/2021 (GISAID, https://www.gisaid.org; accession nos. EPI4858549, EPI1858551) clustered with G9/BJ94 lineage viruses from southern China from 2018. Neuraminidase genes (accession nos. EPI4858550, EPI1858552) clustered with G9/BJ94 lineage viruses from Laos from 2019. Overall, genetic distances between human and chicken viruses support a possible recent shared ancestor consistent with household chickens as the source of exposure.
Seroprevalence studies in rural Cambodia populations show neutralizing antibodies against H9N2 viruses of 1.1%–2.6%, indicative of undetected infections (9,10: Appendix 1 reference 13), similar to avian-exposed persons elsewhere (Appendix 1 reference 14). Therefore, the true burden of human H9N2 virus infections is likely higher than observed. Several human infections with H9N2 were reported in 2020, coinciding with the coronavirus pandemic (Appendix 1 reference 15), when global human seasonal influenza declined, suggesting that sentinel surveillance systems continued to detect seasonal and zoonotic viruses (Appendix 1 reference 16).
Endemic H9N2 and other AIVs in poultry remain a concern for zoonotic infection. An interdisciplinary One Health approach is warranted to curb continuing expansion and emergence. Early detection and control in animal populations, enhanced biosafety, candidate vaccines, and prompt antiviral treatment might mitigate the risks of reassortment and continued evolution that could result in H9N2 viruses with increased mammalian adaptation or human-to-human transmission potential.
Mr. Um is a technical officer in Influenza Surveillance and Response with the National Institute of Public Health and the Centers for Disease Control in the Ministry of Health, Phnom Penh, Cambodia; his research interests include the national influenza surveillance system in Cambodia. Mr. Siegers is a postdoctoral fellow in the Virology Unit at Institute Pasteur du Cambodge in Phnom Penh, Cambodia; his research interests include emerging and endemic infectious diseases in Southeast Asia, especially concerning spillover and exposure to avian influenza viruses.
Acknowledgments
Sequencing primer sequences were generously provided by Peter Thielen. We thank everyone involved in the investigation, including teams at the National Institute for Public Health/National Public Health Laboratory, Centers for Disease Control in Cambodia, Ministry of Health, US Centers for Disease Control and Prevention, and the Influenza Team in the Virology Unit at Institut Pasteur du Cambodge who contributed to this study. We thank all those involved in Influenza Sentinel Surveillance in Cambodia, especially Angkor Children’s Hospital, Siem Reap. We thank the field teams from the National Animal Health and Production, especially Bun Chan and Chhim Vutha. We also thank all of the support teams, including the drivers and facilities personnel who made these studies possible, as well as all the local teams, epidemiologists, veterinary officers, and other staff from the Provincial Health Department in Siem Reap, the Provincial Office of Animal Health and Production, and the Applied Veterinary Epidemiology Training program, Cambodia, especially Sim Thorath, Phy Thorath, and Prum Vich. Finally, we thank all researchers who have submitted sequences to GenBank and GISAID (Appendix 2).
This investigation was undertaken as part of a public health outbreak response and was considered a nonresearch investigation by the Cambodia Ministry of Health. Work at Institut Pasteur in Cambodia was supported by the Office of the Assistant Secretary for Preparedness and Response, US Department of Health and Human Services (grant no. IDSEP 140020-01-00; http://www.asideproject.org), and by the Food and Agriculture Organization, with funding from the United States Agency for International Development under the Emerging Pandemics Threats 2 project.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the Food and Agriculture Organization of the United Nations.
References
- Carnaccini S, Perez DR. H9 Influenza Viruses: An Emerging Challenge. Cold Spring Harb Perspect Med. 2020;10:
a038588 . DOIPubMedGoogle Scholar - Group SHW; SJCEIRS H9 Working Group. Assessing the fitness of distinct clades of influenza A (H9N2) viruses. Emerg Microbes Infect. 2013;2:
e75 .PubMedGoogle Scholar - Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, et al. Human infection with influenza H9N2. Lancet. 1999;354:916–7. DOIPubMedGoogle Scholar
- World Health Organization. Antigenic and genetic characteristics of zoonotic influenza A viruses and development of candidate vaccine viruses for pandemic preparedness; 2021. [cited 2021 March 30]; https://www.who.int/influenza/vaccines/virus/202103_zoonotic_vaccinevirusupdate.pdf
- Pu J, Yin Y, Liu J, Wang X, Zhou Y, Wang Z, et al. Reassortment with dominant chicken H9N2 influenza virus contributed to the fifth H7N9 virus human epidemic. J Virol. 2021;JVI.01578-20.
- Centers for Disease Control and Prevention. Summary of Influenza Risk Assessment Tool (IRAT) results. 2021 [cited 2021 March 30]. https://www.cdc.gov/flu/pandemic-resources/monitoring/irat-virus-summaries.htm
- Karlsson EA, Horm SV, Tok S, Tum S, Kalpravidh W, Claes F, et al. Avian influenza virus detection, temporality and co-infection in poultry in Cambodian border provinces, 2017-2018. Emerg Microbes Infect. 2019;8:637–9. DOIPubMedGoogle Scholar
- Suttie A, Tok S, Yann S, Keo P, Horm SV, Roe M, et al. The evolution and genetic diversity of avian influenza A(H9N2) viruses in Cambodia, 2015 - 2016. PLoS One. 2019;14:
e0225428 . DOIPubMedGoogle Scholar - Blair PJ, Putnam SD, Krueger WS, Chum C, Wierzba TF, Heil GL, et al. Evidence for avian H9N2 influenza virus infections among rural villagers in Cambodia. J Infect Public Health. 2013;6:69–79. DOIPubMedGoogle Scholar
- Horm SV, Tarantola A, Rith S, Ly S, Gambaretti J, Duong V, et al. Intense circulation of A/H5N1 and other avian influenza viruses in Cambodian live-bird markets with serological evidence of sub-clinical human infections. Emerg Microbes Infect. 2016;5:
e70 . DOIPubMedGoogle Scholar
Figure
Cite This ArticleOriginal Publication Date: September 08, 2021
1These authors contributed equally to this article.
Table of Contents – Volume 27, Number 10—October 2021
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Erik A. Karlsson, Virology Unit, Institute Pasteur du Cambodge, 5 Monivong Blvd, PO Box 983, Phnom Penh, Cambodia
Top