Seoul Virus Associated with Pet Rats, Scotland, UK, 2019
James G. Shepherd, Andrew E. Blunsum, Stephen Carmichael, Katherine Smollett, Hector Maxwell-Scott, Eoghan C.W. Farmer, Jane Osborne, Alasdair MacLean, Shirin Ashraf, Rajiv Shah, Rory Gunson, Ana da Silva Filipe, Emma J. Aarons, and Emma C. Thomson

Author affiliations: MRC–University of Glasgow Centre for Virus Research, Glasgow, Scotland, UK (J.G. Shepherd, S. Carmichael, K. Smollett, S. Ashraf, R. Shah, A. da Silva Filipe, E.C. Thomson); National Health Service Greater Glasgow and Clyde, Glasgow (A.E. Blunsum, E.C.W. Farmer); Public Health England, London, UK (H. Maxwell-Scott, J. Osborne, E.J. Aarons); West of Scotland Specialist Virology Centre, Glasgow (A. MacLean, R. Gunson); London School of Hygiene and Tropical Medicine, London (E.C. Thomson)
Main Article
Figure
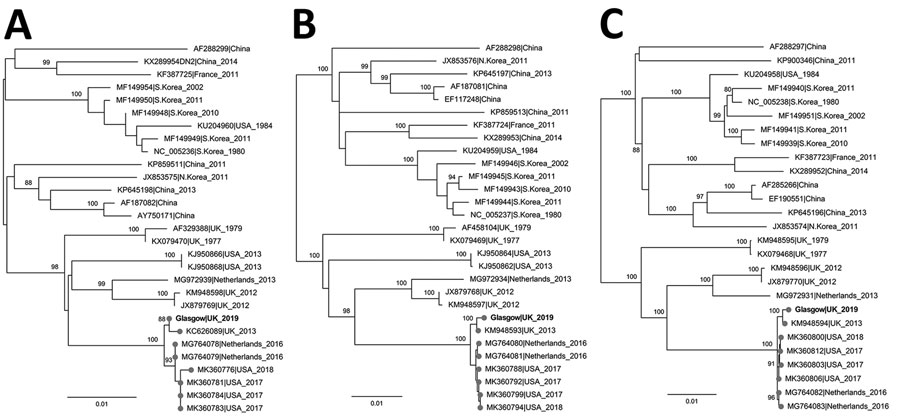
Figure. Maximum-likelihood phylogenetic tree based on the small (A), medium (B), and large (C) segments of Seoul virus isolated from a patient in Scotland, United Kingdom, 2020 (bold text; GenBank accession nos. MZ343375–7), and reference sequences. Isolate names indicate GenBank accession number as well as location and date of isolate. Phylogenetic relationships inferred by RAxML (https://github.com/stamatak/standard-RAxML) using the general time-reversible plus gamma distribution plus invariable site model as determined by jModeltest (https://github.com/ddarriba/jmodeltest2). The tree was rooted at midpoint. Numbers to the left of nodes indicate bootstrap values based on 1,000 replicates. Gray circles indicate sequences associated with domesticated rats. Scale bar indicates substitutions per site.
Main Article
Page created: July 20, 2021
Page updated: September 19, 2021
Page reviewed: September 19, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
