Volume 27, Number 11—November 2021
Dispatch
Co-infection with Legionella and SARS-CoV-2, France, March 2020
Figure 2
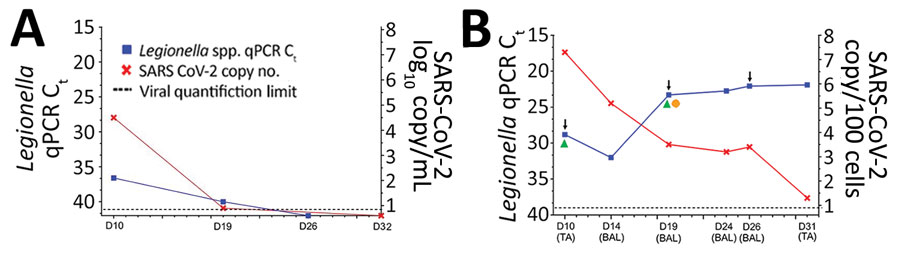
Figure 2. Timeline of detection of SARS-CoV-2 and Legionella in patient 1, a 71-year-old man receiving chemotherapy for multiple myeloma, France, March 2020. A) Serum Legionella DNA load estimated by qPCR Ct and SARS-CoV-2 RNA load expressed as the number of log10 RNA copies/mL serum. B) Pulmonary Legionella DNA load estimated by qPCR Ct, targeting the 16sRNA gene (R-DiaLeg, Diagenode, https://www.diagenode.com) and SARS-CoV-2 RNA load (Institut Pasteur, Paris protocol). Arrows indicate dates of computed tomography scans, green triangles indicates dates of positive Legionella cultures, and orange circle indicates date of lung microbiome testing. Values are normalized according to cellular quantification using the CELL Control r-gene kit (bioMérieux, https://www.biomerieux.com) and expressed as the number of log10 RNA copies/100 cells from pulmonary TA and BAL. BAL, broncho-alveolar lavage; Ct, cycle threshold; D, days after onset of symptoms; qPCR, quantitative PCR; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TA, tracheal aspirations.
1Members are listed at the end of this article.