Volume 27, Number 11—November 2021
Research Letter
Nonclonal Burkholderia pseudomallei Population in Melioidosis Case Cluster, Sri Lanka
Cite This Article
Citation for Media
Abstract
A melioidosis case cluster of 10 blood culture–positive patients occurred in eastern Sri Lanka after an extreme weather event. Four infections were caused by Burkholderia pseudomallei isolates of sequence type 594. Whole-genome analysis showed that the isolates were genetically diverse and the case cluster was nonclonal.
Melioidosis, an emerging tropical infection caused by the soil bacterium Burkholderia pseudomallei, is found most commonly in northern Australia and the tropical countries of Southeast Asia. Melioidosis is endemic in Sri Lanka and has a case-fatality rate of 24% (1). The primary route of acquisition is inoculation of contaminated surface water or soil through skin and mucous membranes. However, a high incidence of pneumonia with sepsis has been reported during extreme weather events, such as heavy rainfall, indicating that inhalation of aerosolized bacteria during cyclones and typhoons is a likely mode of transmission in this setting (2).
We identified a case cluster of 10 blood culture–positive cases of melioidosis in the Batticaloa District of the Eastern Province of Sri Lanka during November–December 2015, after a flooding event; 4 case-patients died (Appendix Table). Before this case cluster was identified, 1 case had been reported from Batticaloa in March 2015, and after this case cluster, 5 cases were found in 2016. We confirmed isolates as B. pseudomallei by real-time PCR to detect the lpxo gene (3). We identified 6 sequence types (STs) by multilocus sequence typing (MLST) (4); of the 10 isolates, 4 were ST594, belonging to the uncommon B. thailandensis–like flagellum and chemotaxis (BTFC) gene cluster. Although the monthly average rainfall for October in the Batticaloa District is usually 160 mm, ≈331.5 mm of rain was recorded in the 48 hours ending at 8:30 a.m. Previous evidence has shown that, in this setting, infections are usually caused by diverse strains because of widespread aerosolization of multiple clones (2). We performed whole-genome sequencing (WGS) to determine clonality among the isolates of identical ST. The Ethics Review Committee of the Faculty of Medicine, University of Colombo, (Colombo, Sri Lanka), approved this study.
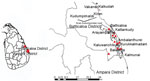
We mapped the geographic location of the cases on Google Earth using ArcGIS version 10.1 (ESRI, https://www.esri.com) (Figure). We compared WGS data of 3 ST594 B. pseudomallei isolates (114, 122, and 133) using binary alignment files with Unipro UGENE version 33 (5). We determined the distribution of genomic islands (GIs) using the Island Viewer 4 web tool (6) and the National Center for Biotechnology Information Multiple Sequence Alignment (https://www.ncbi.nlm.nih.gov/tools/msaviewer) and BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) tools. We used the PHAge Search Tool Enhanced Release (PHASTER) webserver to analyze the annotated bacterial genomes to locate prophage-specifying DNA regions (7). We manually searched the sequences identified by PHASTER and used the position of integrase and last phage–related genes to determine genome boundaries (8). We predicted and annotated all open reading frames (ORFs) of the prophages by PHASTER and BLASTp. We also determined the intracellular motility factor (bimA) allele type present in each strain as previously described (9).
WGS showed genetic diversity in terms of gene content, location, number and type of GIs, prophage DNA distribution, number of intact prophages and bimA allele type among the isolates of ST594 (Table). These results show that the case cluster was nonclonal in origin.
This case cluster after heavy rainfall and flooding in Sri Lanka confirmed that B. pseudomallei is present in the environment of eastern Sri Lanka. The case-fatality rate of the cluster (40%) was almost double that of sporadic infections in Sri Lanka (23%) (1). Six of the 10 isolates in our study belonged to diverse STs; 4 isolates were novel (ST1364, ST1442, ST1179, and ST1413). However, 4 isolates belonging to ST594 were from patients from different geographic locations. In addition, WGS analysis of prophage DNA distribution was able to discriminate between the isolates. GI variation in B. pseudomallei is known to be associated with genomic plasticity of the organism, and GIs appear to be a main source of genomic diversity within B. pseudomallei that can be useful in identifying genetically diverse strains (10). The strain-specific variations in the structure and distribution of GIs in the isolates in this case cluster indicate the presence of genetically diverse strains.
As weather patterns change globally and include more severe weather events and increased flooding, we may see more such case clusters of melioidosis. Physicians in tropical regions must be vigilant for such occurrences because of the high mortality rate associated with melioidosis in this setting. Primary prevention of melioidosis is difficult because of the saprophytic nature of B. pseudomallei and regular flooding during the rainy season. The predominance of diabetic patients in this cluster (Appendix Table) illustrates the importance of detection and control of diabetes in high-risk communities for primary prevention. Secondary prevention requires close coordination between clinicians and microbiologists to encourage patients to seek medical advice early in illness and to transfer febrile patients promptly to centers with blood culture facilities. Early suspicion of melioidosis will enable timely, effective antimicrobial treatment.
Dr. Jayasinghearachchi is a senior lecturer at General Sir John Kotelawala Defense University, Ratmalana. Her research interests include molecular evolution of human pathogens and their epidemiology.
Acknowledgments
We thank K. Arulmoly, P. Mayurathan, S.A. Jauffar, S. Prasath, V. Jeyathas, and M.T.A. Rikarz, and other clinical staff of the Teaching Hospital, Batticaloa. We thank the staff of the Genetech Research Institute and the Department of Microbiology, Faculty of Medicine, University of Colombo, for laboratory assistance. We thank Sameera Fernando, Anjana Rahubaddha, Rajitha Fernando, Kasun de Soyza, Thanuka Gunawardana, and Shehan Dayarathna for assistance with sample and data collection.
This study was funded by US Army Medical Research Acquisition Activity (grant no. W81XWH14C0071) and partially funded by National Research Council, Sri Lanka (grant no. NRC 18079 to A.D.D.S.).
References
- Corea EM, de Silva AD, Thevanesam V. Melioidosis in Sri Lanka. Trop Med Infect Dis. 2018;3:22. DOIPubMedGoogle Scholar
- Ko WC, Cheung BM, Tang HJ, Shih HI, Lau YJ, Wang LR, et al. Melioidosis outbreak after typhoon, southern Taiwan. Emerg Infect Dis. 2007;13:896–8. DOIPubMedGoogle Scholar
- Merritt A, Inglis TJ, Chidlow G, Harnett G. PCR-based identification of Burkholderia pseudomallei. Rev Inst Med Trop São Paulo. 2006;48:239–44. DOIPubMedGoogle Scholar
- Jayasinghearachchi HS, Corea EM, Krishnananthasivam S, Sathkumara HD, Francis VR, Abeysekere TR, et al. Whole-genome sequences of eight clinical isolates of Burkholderia pseudomallei from melioidosis patients in eastern Sri Lanka. Microbiol Resour Announc. 2019;8:e00645–19. DOIPubMedGoogle Scholar
- Okonechnikov K, Golosova O, Fursov M; UGENE team. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–7. DOIPubMedGoogle Scholar
- Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, et al.; Simon Fraser University Research Computing Group. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45(W1):W30–5. DOIPubMedGoogle Scholar
- Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44(W1):
W16-21 . DOIPubMedGoogle Scholar - Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;15;28(12):1647–9.
- Sitthidet C, Stevens JM, Chantratita N, Currie BJ, Peacock SJ, Korbsrisate S, et al. Prevalence and sequence diversity of a factor required for actin-based motility in natural populations of Burkholderia species. J Clin Microbiol. 2008;46:2418–22. DOIPubMedGoogle Scholar
- Holden MT, Titball RW, Peacock SJ, Cerdeño-Tárraga AM, Atkins T, Crossman LC, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A. 2004;101:14240–5. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: August 11, 2021
Table of Contents – Volume 27, Number 11—November 2021
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Enoka M. Corea, Department of Microbiology, Faculty of Medicine, University of Colombo, Colombo, Sri Lanka
Top