Real-Time Genomic Surveillance for SARS-CoV-2 Variants of Concern, Uruguay
Natalia Rego
1, Alicia Costábile
1, Mercedes Paz
1, Cecilia Salazar
1, Paula Perbolianachis
1, Lucía Spangenberg, Ignacio Ferrés, Rodrigo Arce, Alvaro Fajardo, Mailen Arleo, Tania Possi, Natalia Reyes, Ma Noel Bentancor, Andrés Lizasoain, María José Benítez, Viviana Bortagaray, Ana Moller, Gonzalo Bello, Ighor Arantes, Mariana Brandes, Pablo Smircich, Odhille Chappos, Melissa Duquía, Belén González, Luciana Griffero, Mauricio Méndez, Ma Pía Techera, Juan Zanetti, Bernardina Rivera, Matías Maidana, Martina Alonso, Cecilia Alonso, Julio Medina, Henry Albornoz, Rodney Colina, Veronica Noya, Gregorio Iraola, Tamara Fernández-Calero

, Gonzalo Moratorio

, and Pilar Moreno

Author affiliations: Institut Pasteur, Montevideo, Uruguay (N. Rego, A. Costábile, M. Paz, C. Salazar, P. Perbolianachis, L. Spangenberg, I. Ferres, R. Arce, A. Fajardo, M. Brandes, B. Rivera, M. Maidana, M. Alonso, G. Iraola, T. Fernandez-Calero, G. Moratorio, P. Moreno); Universidad de la República, Montevideo, Uruguay (A. Costabile, P. Perbolianachis, R. Arce, A. Fajardo, A. Lizasoain, M.J. Benítez, V. Bortagaray, A. Moller, O. Chappos, M. Duquía, B. González, L. Griffero, M. Méndez, M.P. Techera, J. Zanetti, C. Alonso, R. Colina, G. Moratorio, P. Moreno); Sanatorio Americano, Montevideo (M. Arleo, T. Possi, N. Reyes, M.N. Bentancor, V. Noya); Instituto Oswaldo Cruz–Fiocruz, Rio de Janeiro, Brazil (G. Bello, I. Arantes); Instituto de Investigaciones Biológicas Clemente Estable, Montevideo (P. Smirnich); Ministerio de Salud Pública, Montevideo (J. Medina, H. Albornoz)
Main Article
Figure
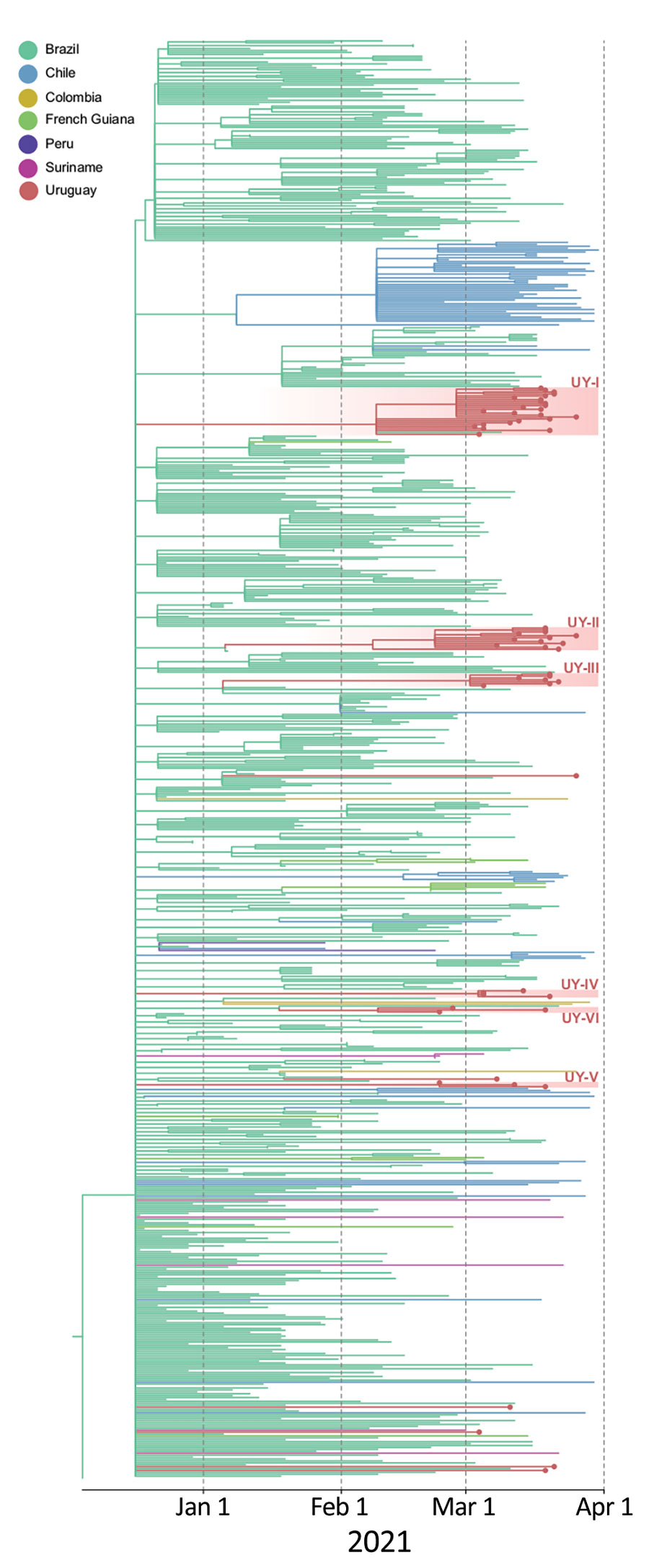
Figure. Time-scaled maximum likelihood Bayesian phylogeographic maximum clade credibility tree of 59 severe acute respiratory syndrome coronavirus 2 lineage P.1 whole-genome sequences from Uruguay and 691 reference sequences from South America. The tree was rooted with the EPI_ISL_833137 sequence from GISAID (https://www.gisaid.org), collected December 4, 2020. Branches are colored according to the most probable location state of their descendant nodes as indicated at the legend. Sequences from Uruguay are shown with dots at the end of the branch. Red shading indicates clades from Uruguay and their distribution along the P.1 tree demonstrates >12 independent introductions and locally transmitted clusters of 3–24 sequences. The tree suggests Brazil has been the source of P.1 dissemination to Uruguay and other countries in South America.
Main Article
Page created: July 27, 2021
Page updated: October 19, 2021
Page reviewed: October 19, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
