Volume 27, Number 11—November 2021
Research Letter
Spontaneous Bacterial Peritonitis Caused by Bordetella hinzii
Figure
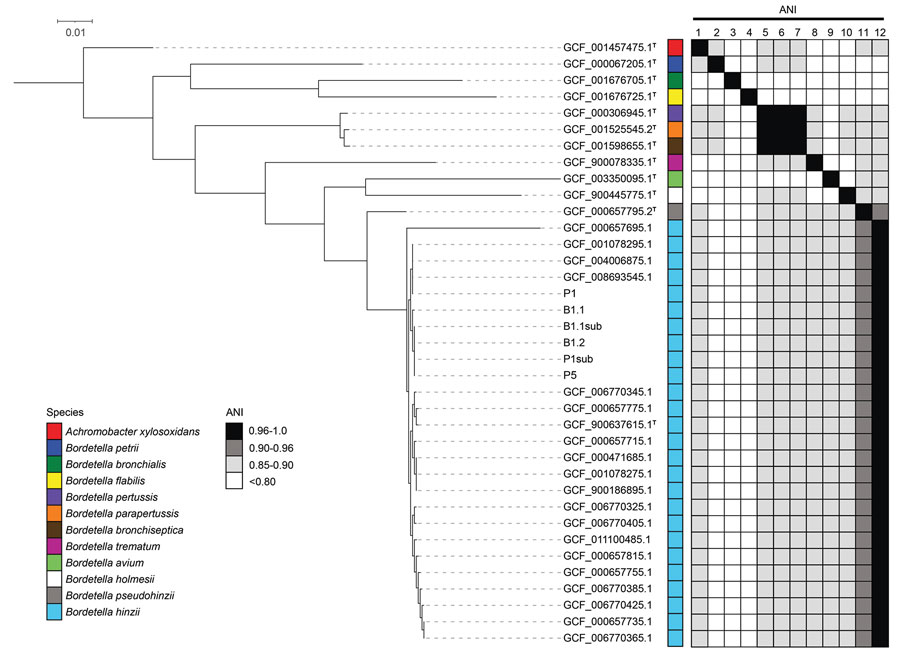
Figure. Comparative genomic analyses of Bordetella hinzii isolates from a patient in Missouri, USA, with type and nontype Bordetella assemblies. After core-genome alignment (58 total core genes), a neighbor-joining phylogenetic tree rooted with Achromobacter xylosoxidans as the outgroup demonstrates the isolates from this study cluster with other previously deposited B. hinzii genomes. Pairwise ANI was performed against type assemblies. The isolates in this study meet the ANI threshold (>0.96%) for species-level identity with B. hinzii type assembly GCF_900637615.1 (7). Isolates were recovered from peritoneal fluid cultures collected at day 1 and day 5 (P1 and P5, respectively; P2sub is a subculture of P1). Blood isolates were recovered from blood cultures collected on day 1 (B1.1 and B1.2; B1.1sub is a subculture of B1.1). As previously observed (8), the type genomes for B. pertussis, B. parapertussis, and B. bronchiseptica represent an instance of previously established, distinct species that exceed the species-level ANI threshold relative to each other. T indicates assemblies generated from type material. Type assemblies are numbered 1–12 on vertical axes as follows: 1, GCF_001457475.1; 2, GCF_000067205.1; 3, GCF_001676705.1; 4, GCF_001676725.1; 5, GCF_000306945.1; 6, GCF_001525545.2; 7, GCF_001598655.1; 8, GCF_900078335.1; 9, GCF_003350095.1; 10, GCF_900445775.1; 11, GCF_000657795.2; 12, GCF_900637615.1. ANI, average nucleotide identity.
References
- Register KB, Kunkle RA. Strain-specific virulence of Bordetella hinzii in poultry. Avian Dis. 2009;53:50–4. DOIPubMedGoogle Scholar
- Jiyipong T, Morand S, Jittapalapong S, Raoult D, Rolain JM. Bordetella hinzii in rodents, Southeast Asia. Emerg Infect Dis. 2013;19:502–3. DOIPubMedGoogle Scholar
- Cookson BT, Vandamme P, Carlson LC, Larson AM, Sheffield JV, Kersters K, et al. Bacteremia caused by a novel Bordetella species, “B. hinzii”. J Clin Microbiol. 1994;32:2569–71. DOIPubMedGoogle Scholar
- Arvand M, Feldhues R, Mieth M, Kraus T, Vandamme P. Chronic cholangitis caused by Bordetella hinzii in a liver transplant recipient. J Clin Microbiol. 2004;42:2335–7. DOIPubMedGoogle Scholar
- Weigand MR, Changayil S, Kulasekarapandian Y, Tondella ML. Complete genome sequences of two Bordetella hinzii strains isolated from humans. Genome Announc. 2015;3:1–2. DOIPubMedGoogle Scholar
- Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–31. DOIPubMedGoogle Scholar
- Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–51. DOIPubMedGoogle Scholar
1These first authors contributed equally to this article.
2Current affiliation: Centers for Disease Control and Prevention, Atlanta, Georgia, USA.
3These authors contributed equally to this article as co–principal investigators.