Volume 27, Number 12—December 2021
Research Letter
Real-Time Projections of SARS-CoV-2 B.1.1.7 Variant in a University Setting, Texas, USA
Figure
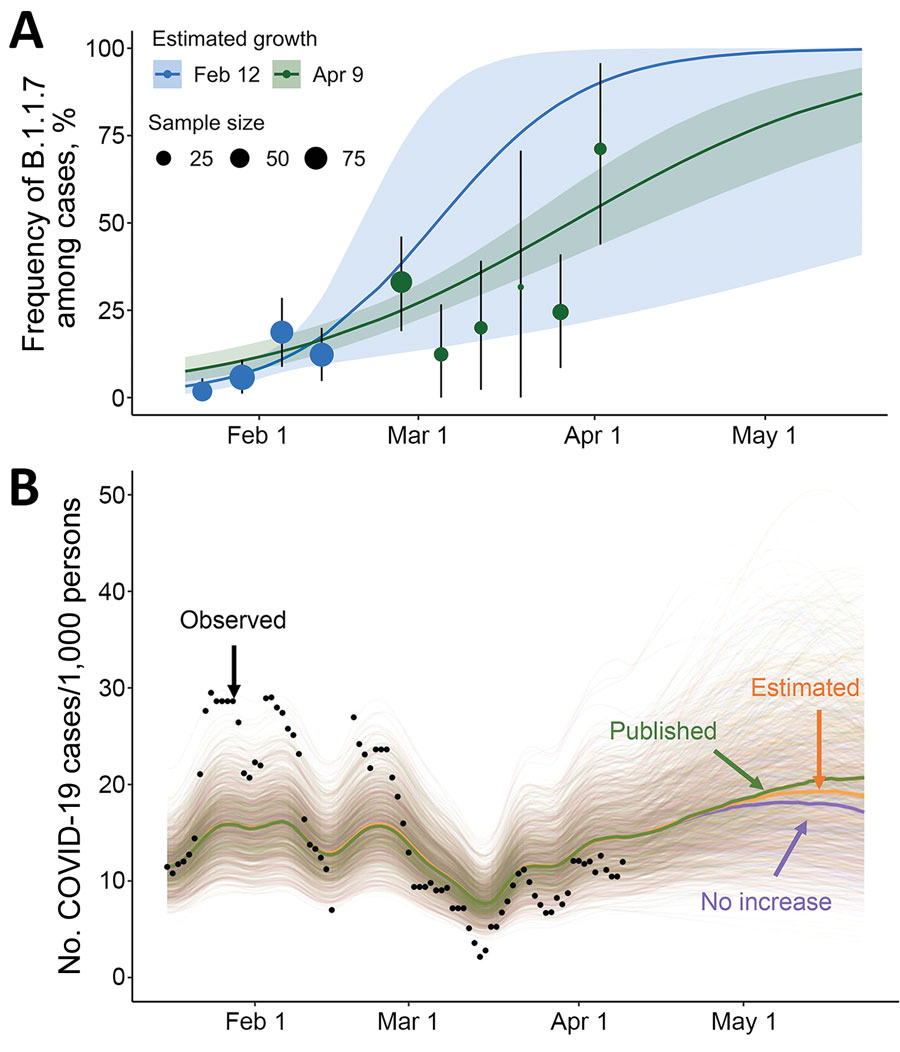
Figure. Estimated frequency of the B.1.1.7 variant among COVID-19 cases at the University of Texas and its projected impact on COVID-19 prevalence, Texas, USA, January 16–May 23, 2021. A) On the basis of the number of samples with spike gene target failures among severe acute respiratory syndrome coronavirus 2–positive samples reported by the University of Texas Proactive Community Testing Program (PCT), we estimated the weekly frequency of the B.1.1.7 variant (points); vertical error bars indicate 95% CIs. We fit a logistic growth model to data through February 12 (blue) and April 9 (green) to project the prevalence of the B.1.1.7 variant relative to the previously circulating wild-type virus through May 23. Shaded bands indicate 95% credible intervals, which reflect uncertainty in the percentage of cases that are spike gene dropouts, the percentage of spike gene dropouts that are B.1.1.7, and the fitted model parameters. The 95% credible interval of our initial projections (blue shading) contains the posterior median estimated from subsequent data (green line). B) Projected COVID-19 cases at the University of Texas through the end of the spring semester. Green, orange, and purple indicate projections with variant transmissibility from published literature, with the university-derived estimate, and with no transmissibility increase from the variant, respectively; black dots indicate the 7-day average reported positive cases per 1,000 persons detected through PCT. The projections assume a reproduction number (Rt) of 1.17 (95% CI 0.94–1.43) as of April 9, on the basis of a recent estimate from PCT data (5,6). Spaghetti lines display 500 simulations; bold lines indicate the median projected value on each day. A lower-transmission scenario is described in the Appendix (https://wwwnc.cdc.gov/EID/article/27/12/21-0652-App1.pdf). COVID-19, coronavirus disease.
References
- Du Z, Wang L, Yang B, Ali ST, Tsang TK, Shan S, et al. Risk for international importations of variant SARS-CoV-2 originating in the United Kingdom. Emerg Infect Dis. 2021;27:1527–9. DOIPubMedGoogle Scholar
- Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, et al.; CMMID COVID-19 Working Group; COVID-19 Genomics UK (COG-UK) Consortium. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:
eabg3055 . DOIPubMedGoogle Scholar - Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A, et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:95–9. DOIPubMedGoogle Scholar
- Washington NL, Gangavarapu K, Zeller M, Bolze A, Cirulli ET, Schiabor Barrett KM, et al. Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell. 2021;184:2587–2594.e7. DOIPubMedGoogle Scholar
- University of Texas at Austin. University of Texas Proactive Community Testing Program for COVID-19 [cited 2021 Feb 9]. https://healthyhorns.utexas.edu/coronavirus_proactive_testing.html
- Cori A, Ferguson NM, Fraser C, Cauchemez S. A new framework and software to estimate time-varying reproduction numbers during epidemics. Am J Epidemiol. 2013;178:1505–12. DOIPubMedGoogle Scholar
- Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, et al.; COVID-19 Genomics UK (COG-UK) consortium. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–9. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.