Volume 27, Number 12—December 2021
Dispatch
Incidence Trends for SARS-CoV-2 Alpha and Beta Variants, Finland, Spring 2021
Figure 2
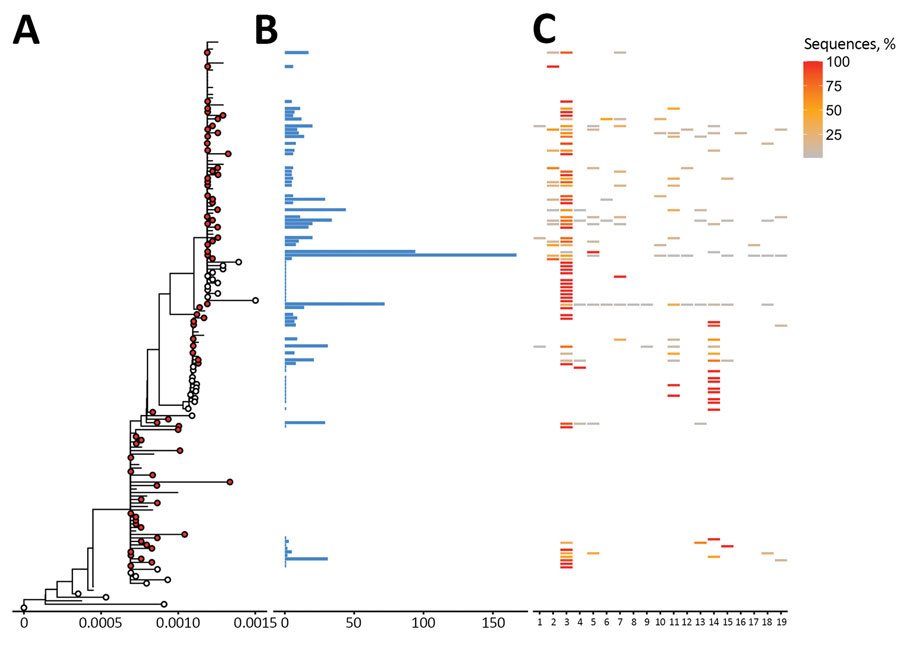
Figure 2. Phylogenetic trees of severe acute respiratory syndrome coronavirus 2 Beta (B1.351) variant clusters from Finland and sequence distribution. The tree (A) shows 76 clusters with ≥5 sequences (red circles), of which 48 contain 898 sequences sampled in Finland using TreeCluster (11), and 23 Finland singletons (white circles) from 33. The tree was constructed by using IQ-TREE 2 (10) with 1,000 ultrafast bootstraps. Each row in subsequent panels is equivalent to a cluster and shows the number of sequences from Finland (B) and the proportion of sequences per region of from Finland (C). Regions of Finland: 1, Central Finland Health Care District; 2, East Savo Hospital District; 3, Hospital District of Helsinki and Uusimaa; 4, Hospital District of South Ostrobothnia; 5, Hospital District of Southwest Finland; 6, Kainuu Social and Health Care Joint Authority; 7, Kanta-Häme Hospital District; 8, Länsi-Pohja Healthcare District; 9, Lapland Hospital District; 10, North Karelia Hospital District; 11, North Ostrobothnia Hospital District; 12, North Savo Hospital District; 13, Päijät-Häme Hospital District; 14, Pirkanmaa Hospital District; 15, Satakunta Hospital District; 16, Social and Health Services in Kymenlaakso; 17, South Karelia Social and Health Care District; 18, South Savo Hospital District; 19, Vaasa Hospital District,
References
- Public Health England. Investigation of SARS-CoV-2 variants of concern [cited 2021 Feb 16]. https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201
- Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–43. DOIPubMedGoogle Scholar
- Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DDS, Mishra S, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–21. DOIPubMedGoogle Scholar
- Campbell F, Archer B, Laurenson-Schafer H, Jinnai Y, Konings F, Batra N, et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021;26:
2100509 . DOIPubMedGoogle Scholar - Virtanen J, Uusitalo R, Korhonen EM, Aaltonen K, Smura T, Kuivanen S, et al. Kinetics of neutralizing antibodies of COVID-19 patients tested using clinical D614G, B.1.1.7, and B 1.351 isolates in microneutralization assays. Viruses. 2021;13:996. DOIPubMedGoogle Scholar
- Jalkanen P, Kolehmainen P, Häkkinen HK, Huttunen M, Tähtinen PA, Lundberg R, et al. COVID-19 mRNA vaccine induced antibody responses against three SARS-CoV-2 variants. Nat Commun. 2021;12:3991. DOIPubMedGoogle Scholar
- Davies NG, Jarvis CI, Edmunds WJ, Jewell NP, Diaz-Ordaz K, Keogh RH; CMMID COVID-19 Working Group. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593:270–4. DOIPubMedGoogle Scholar
- O’Toole A, Scher E, Underwood A, Jackson B, Hill V, McCrone JT, et al. Assignment of epidemiological lineages in an emerging pandemic using the Pangolin tool. Virus Evol. 2021;7:veab064.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. DOIPubMedGoogle Scholar
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. [Erratum in Mol Biol Evol. 2020;37:2461]. Mol Biol Evol. 2020;37:1530–4. DOIPubMedGoogle Scholar
- Balaban M, Moshiri N, Mai U, Jia X, Mirarab S. TreeCluster: Clustering biological sequences using phylogenetic trees. PLoS One. 2019;14:
e0221068 . DOIPubMedGoogle Scholar - Finnish Institute for Health and Welfare (THL). COVID-19 cases in the infectious diseases registry [cited 2021 Feb 17]. https://sampo.thl.fi/pivot/prod/en/epirapo/covid19case/fact_epirapo_covid19case
- Finnish Institute for Health and Welfare (THL). COVID-19 vaccinations in Finland: vaccinations over time in hospital care districts per age group [cited 2021 Jul 12]. https://sampo.thl.fi/pivot/prod/en/vaccreg/cov19cov/summary_cov19covareatime
- Finnish Institute for Health and Welfare (THL). Report of the population serology survey of the coronavirus epidemic [Finnish] [cited 2021 Jun 30]. https://www.thl.fi/roko/cov-vaestoserologia/sero_report_weekly.html
- European Centre for Disease Prevention and Control. SARS-CoV-2 variants of concern as of 15 July 2021 [cited 2021 Jul 16]. https://www.ecdc.europa.eu/en/covid-19/variants-concern
1These authors contributed equally to this article.