Volume 27, Number 2—February 2021
Research
Highly Pathogenic Avian Influenza A(H5N8) Virus Spread by Short- and Long-Range Transmission, France, 2016–17
Figure 1
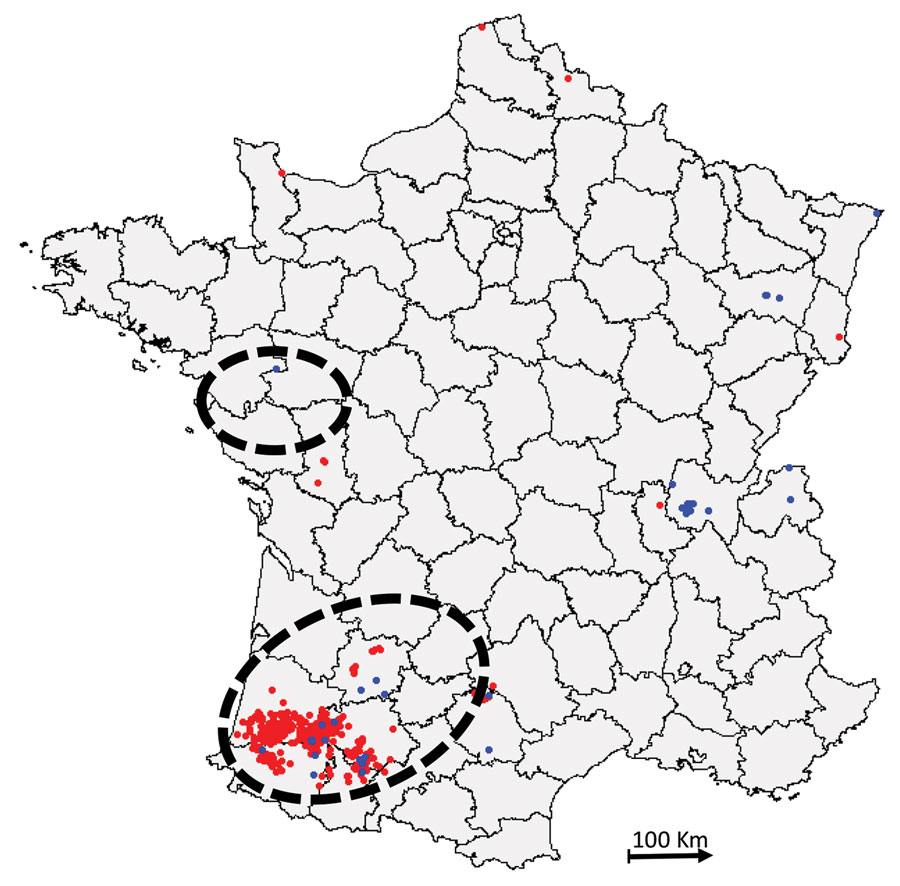
Figure 1. Distribution of highly pathogenic avian influenza H5N8 cases, France, 2016–17 (database of the French National Reference Laboratory for Avian Influenza). Blue indicates cases in wild birds; red indicates cases in domestic or captive birds. Dashed circles indicate zones of high duck farm density (34).
References
- Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus AD, Fouchier RA. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–8. DOIPubMedGoogle Scholar
- Nao N, Yamagishi J, Miyamoto H, Igarashi M, Manzoor R, Ohnuma A, et al. Genetic predisposition to acquire a polybasic cleavage site for highly pathogenic avian influenza virus hemagglutinin. MBio. 2017;8:e02298–16. DOIPubMedGoogle Scholar
- OFFLU. Influenza A cleavage sites. 2020 Jul 8 [cited 2020 Nov 30]. http://www.offlu.net/fileadmin/home/en/resource-centre/pdf/Influenza_A_Cleavage_Sites.pdf
- Pantin-Jackwood MJ, Swayne DE. Pathogenesis and pathobiology of avian influenza virus infection in birds. Rev Sci Tech. 2009;28:113–36. DOIPubMedGoogle Scholar
- Lee DH, Criado MF, Swayne DE. Pathobiological origins and evolutionary history of highly pathogenic avian influenza viruses. Cold Spring Harb Perspect Med. 2020 Jan 21 [Epub ahead of print].
- European Commission. Council directive 2005/94/EC on community measures for the control of avian influenza and repealing directive 92/40/EEC. 2019 Jan 1 [cited 2020 Nov 30]. https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A32005L0094
- WHO/OIE/FAO H5N1 Evolution Working Group. Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1). Emerg Infect Dis. 2008;14:
e1 . DOIPubMedGoogle Scholar - Fusaro A, Zecchin B, Vrancken B, Abolnik C, Ademun R, Alassane A, et al. Disentangling the role of Africa in the global spread of H5 highly pathogenic avian influenza. Nat Commun. 2019;10:5310. DOIPubMedGoogle Scholar
- Saito T, Tanikawa T, Uchida Y, Takemae N, Kanehira K, Tsunekuni R. Intracontinental and intercontinental dissemination of Asian H5 highly pathogenic avian influenza virus (clade 2.3.4.4) in the winter of 2014-2015. Rev Med Virol. 2015;25:388–405. DOIPubMedGoogle Scholar
- Lee DH, Torchetti MK, Winker K, Ip HS, Song CS, Swayne DE. Intercontinental spread of Asian-origin H5N8 to North America through Beringia by migratory birds. J Virol. 2015;89:6521–4. DOIPubMedGoogle Scholar
- Lee DH, Bertran K, Kwon JH, Swayne DE. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J Vet Sci. 2017;18(S1):269–80. DOIPubMedGoogle Scholar
- Cattoli G, Fusaro A, Monne I, Capua I. H5N1 virus evolution in Europe—an updated overview. Viruses. 2009;1:1351–63. DOIPubMedGoogle Scholar
- Global Consortium for H5N8 and Related Influenza Viruses. Role for migratory wild birds in the global spread of avian influenza H5N8. Science. 2016;354:213–7. DOIPubMedGoogle Scholar
- King J, Schulze C, Engelhardt A, Hlinak A, Lennermann SL, Rigbers K, et al. Novel HPAIV H5N8 reassortant (clade 2.3.4.4b) detected in Germany. Viruses. 2020;12:281. DOIPubMedGoogle Scholar
- Hill NJ, Hussein IT, Davis KR, Ma EJ, Spivey TJ, Ramey AM, et al. Reassortment of influenza A viruses in wild birds in Alaska before H5 clade 2.3.4.4 outbreaks. Emerg Infect Dis. 2017;23:654–7. DOIPubMedGoogle Scholar
- Antigua KJC, Choi WS, Baek YH, Song MS. The emergence and decennary distribution of clade 2.3.4.4 HPAI H5Nx. Microorganisms. 2019;7:156. DOIPubMedGoogle Scholar
- Pohlmann A, Starick E, Grund C, Höper D, Strebelow G, Globig A, et al. Swarm incursions of reassortants of highly pathogenic avian influenza virus strains H5N8 and H5N5, clade 2.3.4.4b, Germany, winter 2016/17. Sci Rep. 2018;8:15. DOIPubMedGoogle Scholar
- Brown I, Kuiken T, Mulatti P, Smietanka K, Staubach C, Stroud D, et al.; European Food Safety Authority; European Centre for Disease Prevention and Control; European Union Reference Laboratory for Avian influenza. Avian influenza overview September - November 2017. EFSA J. 2017;15:
e05141 .PubMedGoogle Scholar - Lycett SJ, Pohlmann A, Staubach C, Caliendo V, Woolhouse M, Beer M, et al.; Global Consortium for H5N8 and Related Influenza Viruses. Genesis and spread of multiple reassortants during the 2016/2017 H5 avian influenza epidemic in Eurasia. Proc Natl Acad Sci U S A. 2020;117:20814–25. DOIPubMedGoogle Scholar
- Guinat C, Nicolas G, Vergne T, Bronner A, Durand B, Courcoul A, et al. Spatio-temporal patterns of highly pathogenic avian influenza virus subtype H5N8 spread, France, 2016 to 2017. Euro Surveill. 2018;23.
- Bronner A, Niqueux E, Schmitz A, Le Bouquin S, Huneau-Salaûn A, Guinat C, et al. Description of the highly pathogenic avian influenza episode in France in 2016–2017 [in French]. Bulletin épidémiologique, santé animale et alimentation. 2017;79:13–7.
- Slomka MJ, Coward VJ, Banks J, Löndt BZ, Brown IH, Voermans J, et al. Identification of sensitive and specific avian influenza polymerase chain reaction methods through blind ring trials organized in the European Union. Avian Dis. 2007;51(Suppl):227–34. DOIPubMedGoogle Scholar
- Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, et al. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol. 2009;83:10309–13. DOIPubMedGoogle Scholar
- Briand FX, Schmitz A, Ogor K, Le Prioux A, Guillou-Cloarec C, Guillemoto C, et al. Emerging highly pathogenic H5 avian influenza viruses in France during winter 2015/16: phylogenetic analyses and markers for zoonotic potential. Euro Surveill. 2017;22:30473. DOIPubMedGoogle Scholar
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. DOIPubMedGoogle Scholar
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. DOIPubMedGoogle Scholar
- Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003.
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. DOIPubMedGoogle Scholar
- Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol. 2012;29:2157–67. DOIPubMedGoogle Scholar
- Hill V, Baele G. Bayesian estimation of past population dynamics in BEAST 1.10 using the Skygrid coalescent model. Mol Biol Evol. 2019;36:msz172.
- Lanfear R, Hua X, Warren DL. Estimating the effective sample size of tree topologies from Bayesian phylogenetic analyses. Genome Biol Evol. 2016;8:2319–32. DOIPubMedGoogle Scholar
- Vaughan TG. IcyTree: rapid browser-based visualization for phylogenetic trees and networks. Bioinformatics. 2017;33:2392–4. DOIPubMedGoogle Scholar
- Leigh J, Bryant D. POPART: full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–6. DOIGoogle Scholar
- Agence nationale de sécurité sanitaire de l’alimentation, de l’environnement et du travail. Assessing avian influenza risk levels and their evolution [in French]. 2017 [cited 2017 Nov 07]. https://www.anses.fr/fr/system/files/SABA2017SA0203-partie2.pdf
- Fusaro A, Monne I, Mulatti P, Zecchin B, Bonfanti L, Ormelli S, et al. Genetic diversity of highly pathogenic avian influenza A(H5N8/H5N5) viruses in Italy, 2016–17. Emerg Infect Dis. 2017;23:1543–7. DOIPubMedGoogle Scholar
- Świętoń E, Śmietanka K. Phylogenetic and molecular analysis of highly pathogenic avian influenza H5N8 and H5N5 viruses detected in Poland in 2016-2017. Transbound Emerg Dis. 2018;65:1664–70. DOIPubMedGoogle Scholar
- Savić V. Novel reassortant clade 2.3.4.4 avian influenza A(H5N5) virus in wild birds and poultry, Croatia, 2016–2017. Vet Arh. 2017;87:377–96. DOIGoogle Scholar
- Shapiro B, Rambaut A, Drummond AJ. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol. 2006;23:7–9. DOIPubMedGoogle Scholar
- Souvestre M, Guinat C, Niqueux E, Robertet L, Croville G, Paul M, et al. Role of backyard flocks in transmission dynamics of highly pathogenic avian influenza A(H5N8) clade 2.3.4.4, France, 2016–2017. Emerg Infect Dis. 2019;25:551–4. DOIPubMedGoogle Scholar
- Moisson M, Hamon M, Malhere C, Donguy M, Niqueux E, Scoizec A, et al. The situation of HP H5N8 avian influenza in France as of 05/12/2016. 2016 [cited 2016 Dec 06]. https://www.plateforme-esa.fr/article/situation-de-l-influenza-aviaire-hp-h5n8-en-france-au-05122016
- Andronico A, Courcoul A, Bronner A, Scoizec A, Lebouquin-Leneveu S, Guinat C, et al. Highly pathogenic avian influenza H5N8 in south-west France 2016-2017: A modeling study of control strategies. Epidemics. 2019;28:
100340 . DOIPubMedGoogle Scholar - Guinat C, Artois J, Bronner A, Guérin JL, Gilbert M, Paul MC. Duck production systems and highly pathogenic avian influenza H5N8 in France, 2016-2017. Sci Rep. 2019;9:6177. DOIPubMedGoogle Scholar
Page created: December 07, 2020
Page updated: January 23, 2021
Page reviewed: January 23, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.