Volume 27, Number 3—March 2021
Dispatch
Human Infection with Eurasian Avian-Like Swine Influenza A(H1N1) Virus, the Netherlands, September 2019
Cite This Article
Citation for Media
Abstract
We report a zoonotic infection of a pig farmer in the Netherlands with a Eurasian avian-like swine influenza A(H1N1) virus that was also detected in the farmed pigs. Both viruses were antigenically and genetically characterized. Continued surveillance of swine influenza A viruses is needed for risk assessment in humans and swine.
Eurasian avian-like swine influenza A(H1N1) viruses (IAVs) are entirely derived from a precursor virus of avian origin (1) and have been enzootic in the swine population in Europe since 1979 and in Asia since 1993. Zoonotic infections with such viruses, which are then termed H1N1 variant (H1N1v) viruses, occur sporadically. Most cases occur in humans who have direct exposure to pigs. Since 1986, several human cases of Eurasian avian-like H1N1 swine IAV have been reported in Europe (2–4) and China (3,5).
These events reflect the possibility of Eurasian avian-like H1N1 swine IAV transmission from swine to humans. In this study, we report an infection with a Eurasian avian-like H1N1 swine IAV in a pig farmer and his pigs in a herd in the Netherlands. We also conducted whole-genome characterization of viruses from the man and the pigs.
On September 18, 2019, acute respiratory disease was observed in a 43-year-old man (farmer) and his 14-week-old fattening pigs and gilts. The pigs of this closed farm showed coughing, anorexia, tachypnea, dyspnea, and lethargy. Two days earlier, a 44-year-old man (animal caretaker) had reported similar symptoms. The sows of this herd (n = 420) were vaccinated against swine IAVs with Respiporc FLU3 vaccine (Ceva, https://www.ceva.com), but the farmer and animal caretaker were not recently vaccinated against human seasonal influenza viruses. Both humans and the pigs recovered completely within 10 days after the first appearance of signs or symptoms. Family members and close contacts of the men did not show development of influenza-like symptoms.
Six days after onset of disease, nasal swab samples were collected from the farmer, the animal caretaker, and 6 symptomatic pigs. Human samples were collected by self-sampling, and informed consent was obtained from the farmer and the animal caretaker. Subsequently, samples were shipped to the Laboratory of Virology, Faculty of Veterinary Medicine, Ghent University (Merelbeke, Belgium).
Upon inoculation into MDCK cells, IAV was isolated from the sample of the farmer and from a pooled sample of the pigs; no virus was isolated from the animal caretaker. Public health authorities in the Netherlands were notified about the H1N1v infection. The human H1N1v isolate was named A/Netherlands/Gent-193/2019, and the swine H1N1 isolate was named A/swine/Netherlands/Gent-193/2019.
Virus neutralization tests with swine antiserum against swine IAVs of the H1N1, H1N2, and H3N2 subtypes showed an antigenic relationship between both newly discovered isolates and Eurasian avian-like H1N1 swine IAVs from 1998 and 2010, as well as the prototype influenza A(H1N1)pdm09 (pH1N1) A/California/04/2009 virus. Serologic cross-reactivity with H1N2 or H3N2 swine IAVs was not observed (Table 1).
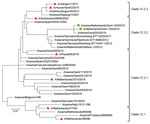
Initial analyses by multiplex real-time reverse transcription PCRs (6) and whole-genome next-generation sequencing (7) of both isolates confirmed that all genome segments were closely related to those of Eurasian avian-like H1N1 swine IAVs. A BLAST homology search (http://www.fludb.org) with both whole genomes showed highest nucleotide identities (96%) for hemagglutinin (HA) and neuraminidase with clade 1C.2.2 Eurasian avian-like H1N1 swine IAVs isolated in Germany and the Netherlands during 2011–2012. These databases contain limited numbers of sequences of this swine IAV clade, which explains the lack of similar recent viruses. A phylogenetic tree of Eurasian avian-like H1N1 swine IAVs isolated in Europe and Asia was constructed by using MEGA7 software (https://www.megasoftware.net). Phylogenetic analysis confirmed the genetic relationship of the HA1 genes of both isolates with Eurasian avian-like H1N1 swine IAVs of clade 1C.2.2 (Table 2; Figure).
The human H1N1v and swine H1N1 isolates differed in several positions in the genes coding for the 3 polymerase proteins (polymerase basic [PB] 2, PB1, and polymerase acidic), the HA gene, and the nonstructural protein gene. The other 3 gene segments (neuraminidase, nucleoprotein, and matrix) were 100% identical. HA gene sequences of the swine H1N1 and the human H1N1v isolates showed amino acid substitutions K142N, N195S, and V215I (H1 numbering). Position 142 is located in antigenic site Ca2 and position 195 in antigenic site Sb (8). In human seasonal influenza A(H1N1) viruses, a substitution at position 142 was reported to cause antigenic change (9). This substitution might explain the loss of reactivity with pH1N1 antiserum for the human H1N1v isolate versus the swine H1N1 isolate (Table 1). In addition, in H5 IAVs this substitution decreased the pH at which the HA underwent fusion (10).
Because human-adapted viruses undergo fusion at a lower pH (5.0–5.5) than swine-adapted and avian-adapted viruses (pH 5.6–6.0), such mutations might contribute to human adaptation of zoonotic viruses. We found multiple substitutions in the polymerase genes of the human H1N1v isolate: R739Q in PB2, L108I and T652A in PB1, and D682N in polymerase acidic. Based on analyses in the FluSurver database (https://flusurver.bii.a-star.edu.sg/), the R739Q substitution in the PB2 gene might influence the binding of PB2 to host protein(s). We also found 2 substitutions in the nonstructural protein gene: V18I and G227R. Sequences were made publicly available in GenBank (accession nos. MT395362–77), and GISAID (https://www.gisaid.org; accession nos. EPI_ISL_430866 [A/swine/Netherlands/Gent-193/2019] and EPI_ISL_0865 [A/Netherlands/Gent-193/2019]).
We report another zoonotic infection with a Eurasian avian-like H1N1 swine IAV in Europe since the emergence of the virus in 1979 (2–4). No further human-to-human transmission was reported, although it cannot be excluded that the farmer was infected by the animal caretaker. The nasal swab sample from the caretaker might have tested negative because it was collected as late as 8 days after he reported influenza-like symptoms.
The swine antiserum against the pH1N1 virus cross-reacted with the swine H1N1 isolate from this investigation (Table 1) but had a 192-fold lower virus neutralization titer against the human H1N1v isolate. Therefore, it is unlikely that current human seasonal vaccines would provide cross-protection against the human H1N1v isolate. This finding is consistent with our recent investigations of human serum samples for antibodies against 8 H1 swine IAVs representing 7 predominant H1 clades of swine IAVs; only 55 (10%) of 549 human serum samples had hemagglutination inhibition titers >40 against a European avian-like H1N1 swine IAV of clade 1C.2.1, which is predominant in swine in Europe (11), compared with 24%–54% against 5 other clades (12). These data point toward a relatively greater zoonotic risk for avian-like H1N1 swine IAVs from Europe and are consistent with previous studies about Eurasian avian-like H1N1 swine IAVs from China (5,13). Our data further support the notion that Eurasian avian-like H1N1 swine IAVs need to be monitored closely.
We found several amino acid substitutions between the H1N1 swine isolate and the H1N1v human isolate, but their role remains obscure. The past 2 decades have seen an unprecedented increase of data for putative mammalian-adaptive mutations of avian influenza viruses. The known genetic markers are mainly based on studies with wholly avian viruses of various HA subtypes in mammalian cell culture or in ferrets. Knowledge of amino acid substitutions that might enable adaptation of swine-adapted influenza viruses to humans, in contrast, is almost nonexistent (14). This finding is true for Eurasian avian-like H1N1 swine IAVs, as well as for the pH1N1 virus, which is the only known swine-origin virus with the ability to spread efficiently between humans. Our study highlights the need for experimental research on this topic and for continued surveillance of swine IAVs because of the risk for human infection or zoonotic spread.
Ms. Parys is a PhD student in the Laboratory of Virology, Faculty of Veterinary Medicine, Ghent University, Merelbeke, Belgium. Her primary research interests are the pig as a model for development of broadly protective influenza A vaccines and public health implications of swine influenza.
Acknowledgments
We thank the farmer, animal caretaker, and veterinarian for their cooperation and Nele Dennequin and Melanie Bauwens for providing technical assistance.
This study was supported by the University of Ghent Research Fund Bijzonder OnderzoeksFonds (grant no. 01J102017) and the European Union Horizon 2020 Research and Innovation Programme (grant no. 727922 DELTA-FLU).
References
- Pensaert M, Ottis K, Vandeputte J, Kaplan MM, Bachmann PA. Evidence for the natural transmission of influenza A virus from wild ducks to swine and its potential importance for man. Bull World Health Organ. 1981;59:75–8.PubMedGoogle Scholar
- Freidl GS, Meijer A, de Bruin E, de Nardi M, Munoz O, Capua I, et al.; FLURISK Consortium. Influenza at the animal-human interface: a review of the literature for virological evidence of human infection with swine or avian influenza viruses other than A(H5N1). Euro Surveill. 2014;19:20793. DOIPubMedGoogle Scholar
- World Health Organization. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness [cited 2020 Jun 18]. https://www.who.int/influenza/vaccines/virus/202009_zoonotic_vaccinevirusupdate.pdf
- European Centre for Disease Prevention and Control. Update: swine-origin triple reassortant influenza A(H3N2) variant viruses in North America, 2012. Stockholm: The Centre [cited 2020 Dec 9]. https://www.ecdc.europa.eu/sites/default/files/media/en/publications/publications/1208-ter-rapid-risk-assessment-influenza-ah3n2-us.pdf
- Sun H, Xiao Y, Liu J, Wang D, Li F, Wang C, et al. Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc Natl Acad Sci U S A. 2020;117:17204–10. DOIPubMedGoogle Scholar
- Henritzi D, Zhao N, Starick E, Simon G, Krog JS, Larsen LE, et al. Rapid detection and subtyping of European swine influenza viruses in porcine clinical samples by haemagglutinin- and neuraminidase-specific tetra- and triplex real-time RT-PCRs. Influenza Other Respir Viruses. 2016;10:504–17. DOIPubMedGoogle Scholar
- King J, Schulze C, Engelhardt A, Hlinak A, Lennermann SL, Rigbers K, et al. Novel HPAIV H5N8 reassortant (Clade 2.3.4.4b) detected in Germany. Viruses. 2020;12:1–7. DOIPubMedGoogle Scholar
- Brownlee GG, Fodor E. The predicted antigenicity of the haemagglutinin of the 1918 Spanish influenza pandemic suggests an avian origin. Philos Trans R Soc Lond B Biol Sci. 2001;356:1871–6. DOIPubMedGoogle Scholar
- Harvey WT, Benton DJ, Gregory V, Hall JPJ, Daniels RS, Bedford T, et al. Identification of low- and high-impact hemagglutinin amino acid substitutions that drive antigenic drift of influenza A (H1N1) viruses. PLoS Pathog. 2016;12:
e1005526 . DOIPubMedGoogle Scholar - Rudneva IA, Timofeeva TA, Ignatieva AV, Shilov AA, Krylov PS, Ilyushina NA, et al. Pleiotropic effects of hemagglutinin amino acid substitutions of H5 influenza escape mutants. Virology. 2013;447:233–9. DOIPubMedGoogle Scholar
- Anderson TK, Macken CA, Lewis NS, Scheuermann RH, Van Reeth K, Brown IH, et al. A phylogeny-based global nomenclature system and automated annotation tool for H1 hemagglutinin genes from swine influenza A viruses. MSphere. 2016;1:e00275–16. DOIPubMedGoogle Scholar
- Vandoorn E, Leroux-Roels I, Leroux-Roels G, Parys A, Vincent A, Van Reeth K. Detection of H1 swine influenza A virus antibodies in human serum samples by age group. Emerg Infect Dis. 2020;26:2118–28. DOIPubMedGoogle Scholar
- Yang H, Chen Y, Qiao C, He X, Zhou H, Sun Y, et al. Prevalence, genetics, and transmissibility in ferrets of Eurasian avian-like H1N1 swine influenza viruses. Proc Natl Acad Sci U S A. 2016;113:392–7. DOIPubMedGoogle Scholar
- Pulit-Penaloza JA, Belser JA, Tumpey TM, Maines TR. Sowing the seeds of a pandemic? Mammalian pathogenicity and transmissibility of H1 variant influenza viruses from the swine reservoir. Trop Med Infect Dis. 2019;4:1–21. DOIPubMedGoogle Scholar
Figure
Tables
Cite This ArticleOriginal Publication Date: February 09, 2021
Table of Contents – Volume 27, Number 3—March 2021
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Kristien Van Reeth, Laboratory of Virology, Faculty of Veterinary Medicine, Ghent University, Salisburylaan 133, 9820 Merelbeke, Belgium
Top