Isolate-Based Surveillance of Bordetella pertussis, Austria, 2018–2020
Adriana Cabal

, Daniela Schmid, Markus Hell, Ali Chakeri, Elisabeth Mustafa-Korninger, Alexandra Wojna, Anna Stöger, Johannes Möst, Eva Leitner, Patrick Hyden, Thomas Rattei, Adele Habington, Ursula Wiedermann, Franz Allerberger, and Werner Ruppitsch
Author affiliations: Institute for Medical Microbiology and Hygiene, Austrian Agency for Health and Food Safety, Vienna, Austria (A. Cabal, D. Schmid, A. Chakeri, A. Stöger, F. Allerberger, W. Ruppitsch); MEDILAB, Teaching Laboratory of the Paracelsus Medical University, Salzburg, Austria (M. Hell, E. Mustafa-Korninger, A. Wojna); Centre for Public Health, Medical University Vienna, Vienna (A. Chakeri); MB-LAB Clinical Microbiology Laboratory, Innsbruck, Austria (J. Möst); Consultant Laboratory for Bordetella of the Robert Koch Institute, Medical University of Graz, Graz, Austria (E. Leitner); Centre for Microbiology and Environmental Systems Science, University of Vienna, Vienna (P. Hyden, T. Rattei); Children’s Health Ireland at Crumlin, Dublin, Ireland (A. Habington); Institute of Specific Prophylaxis and Tropical Medicine, Medical University of Vienna, Vienna (U. Wiedermann)
Main Article
Figure 4
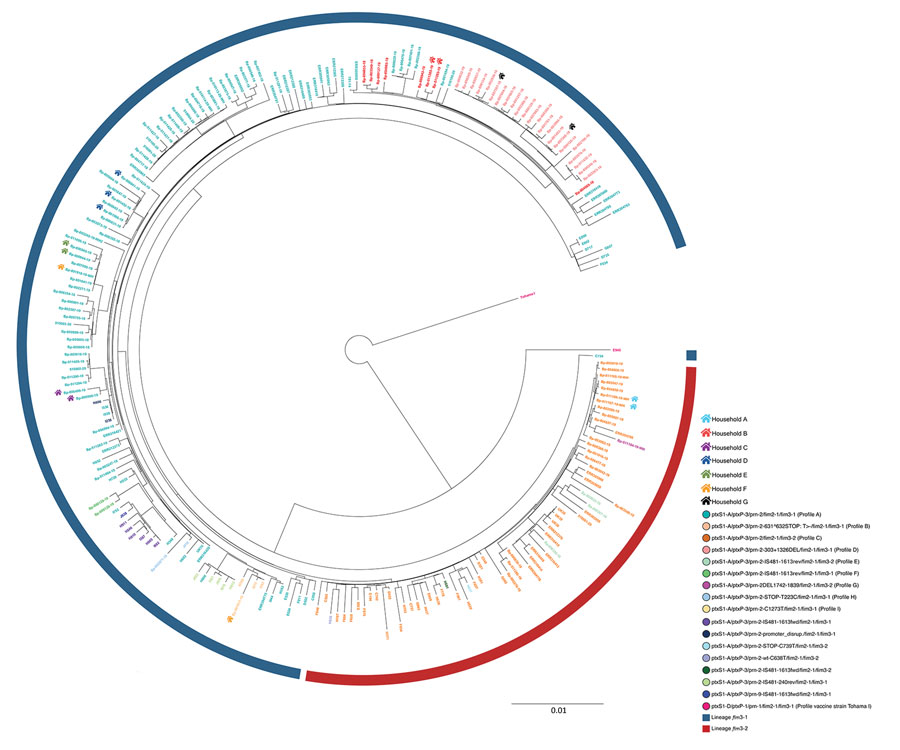
Figure 4. Maximum-likelihood phylogenetic tree generated using core-genome multilocus sequence typing data from 106 outbreak genome sequences from the United States and the United Kingdom and the 123 Bordetella pertussis isolates identified in an isolate-based surveillance study, Austria, May 2018–May 2020. Isolate identifiers are colored by genetic profile. These genetic profiles include profiles A–I, defined in this study, and other genetic profiles described outside of Austria. The circular blue line represents isolates of fim3-1 lineage; the circular red line represents fim3-1 isolates. A color-coded house-like symbol indicates isolates obtained from case-patients living in the same household. Scale bar indicates nucleotide substitutions per site.
Main Article
Page created: January 12, 2021
Page updated: February 21, 2021
Page reviewed: February 21, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
