Volume 27, Number 3—March 2021
Research Letter
Drug-Resistant Tuberculosis in Pet Ring-Tailed Lemur, Madagascar
Cite This Article
Citation for Media
Abstract
We diagnosed tuberculosis in an illegally wild-captured pet ring-tailed lemur manifesting lethargy, anorexia, and cervical lymphadenopathy. Whole-genome sequencing confirmed the Mycobacterium tuberculosis isolate belonged to lineage 3 and harbored streptomycin resistance. We recommend reverse zoonosis prevention and determination of whether lemurs are able to maintain M. tuberculosis infection.
Tuberculosis (TB) is an ancient disease affecting a plethora of domestic and wild animals, including humans. In primates, TB can cause severe multisystemic disease. The prevalence of TB in lemurs within Madagascar is unknown; the most recent documented case occurred in 1973 (1). Reverse zoonotic transmission of TB can occur when nonhuman primates are in close contact with humans (1). We report a clinical case and genomic analysis of TB infection in a female subadult ring-tailed lemur (Lemur catta) held at a nongovernmental organization facility in Southwestern Madagascar. The University of San Diego (San Diego, CA, USA) provided ethics authorization (no. IACUC 0619-01).
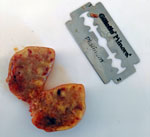
The lemur was born in the wild in September or October 2018 and was surrendered to the facility in April 2019. On July 12, the animal was emaciated, anorexic, and lethargic; it had a large fistulated mass on the left cervical region. The mass was surgically removed and found to be caseous and necrotic (Figure). Despite rehydration and systemic antimicrobial therapy, the lemur died on July 16.
We confirmed TB infection by PCR on the lymph node sample using GeneXpert MTB/RIF assay (Cepheid, https://www.cepheid.com) (2). We cultured on Löwenstein-Jensen solid medium to confirm streptomycin resistance using the proportions method, enabling phenol chloroform DNA extraction and genomic DNA sequencing using Oxford Nanopore Technologies (ONT) (https://www.nanoporetech.com) long-read sequencing. We basecalled raw data using ONT Guppy software version 3.4.5. We performed read mapping using minimap2 version 2.17. For decontamination, we used a manually curated database including viral nontuberculosis mycobacteria and human sequences, augmented with L. catta genome (GenBank accession no. PVHV00000000) to improve host DNA filtering. Decontaminated reads were mapped to the M. tuberculosis H37Rv reference genome (accession no. NC_000962.3); we called single-nucleotide polymorphisms (SNPs) using bcftools version 1.10 (http://samtools.github.io/bcftools/bcftools.html) and masked repetitive regions (3). We performed genotypic resistance testing using Mykrobe Predict version 0.8.2 (https://www.mykrobe.com) and confirmed streptomycin resistance (causative variant R83P/CCG4407954CGG) (4). For lineage identification, we used both SNP-based method, which uses known lineage-defining SNPs, and k-mer-based methods, which rely on an in-silico equivalent of PCR probes analyzing each SNP alleles’ 20bp flanking regions (5–7). Both methods confirmed the isolate as lineage 3.1.1 (Central Asian sublineage Kilimanjaro, CASI-KILI). We ruled out laboratory cross-contamination and assessed relative genomic distance of this isolate compared with other human TB isolates from Madagascar by reanalyzing all lineage 3 TB isolates cultured in the laboratory during March 2017–June 2019 for which lineage typing and genomic sequencing data were available, and created a SNP distance matrix and phylogenetic tree (Appendix Figures 1, 2). The lemur’s isolate was substantially distant from other isolates by a closest SNP distance of 63 SNPs (mada_116), ruling out laboratory contamination or transspecies transmission within the samples processed on site (3). We submitted M. tuberculosis lineage 3 consensus sequence to GenBank (accession no. PRJNA659624).
Human TB isolates in the region of Toliara most frequently belong to lineage 1 (Institut Pasteur de Madagascar, unpub. data). However, lineage 3 isolates were previously isolated in humans from other regions of Madagascar (7). Pet lemurs are transported over vast distances (8,9); this lemur may have originated or been transferred from another region of Madagascar. The Malagasy lineage 3 profile shares similarities with strains found in Tanzania and the Indo-Pakistanese subcontinent (7).
Human activities, including trade and translocation of wild animals and keeping of wildlife as pets, have resulted in reverse zoonotic TB and spillover into wild populations in, among others, meerkats (Suricata suricatta), banded mongoose (Mungos mungo), and Asian elephants (Elephas maximus) (10). Although the risks of transmitting emerging diseases from wildlife to humans have received much attention, the risks that human diseases present to wildlife are not well described. In addition to other anthropogenic activities that imperil wildlife (e.g., deforestation, bushmeat consumption, animal trafficking), the effects of human disease reservoirs may become increasingly detrimental (10).
Illegal trade of wild-captured lemurs is rampant in Madagascar (8,9). Moreover, humans are frequently in close contact with pet or tourist facility–based lemurs. Some resorts encourage tourists to feed lemurs from their mouths, whereby pathogens could be transferred. Because lemurs make poor pets and often become aggressive, many are discarded as adults, some by release into forests with wild conspecific populations (8,9).
We present anatomopathologic and molecular diagnostics evidence that wild-born lemurs can become infected with and die from complications of TB. To minimize risk for transmission of TB between humans and lemurs, we recommend enforced prohibition of keeping wild-captured lemurs as pets, systematic clinical screening and microbiological testing of facility-based animals and staff who become ill, and necropsy of deceased lemurs. Because we do not know if lemurs are able to maintain M. tuberculosis infection, we recommend quarantine and testing at lemur facilities and caution against release of captive lemurs into the wild. We also warn against close proximity or contact between humans and lemurs (captive or wild) in Madagascar, given the potential for reverse zoonotic and zoonotic transmission of TB and other infectious diseases.
Dr. LaFleur is an assistant professor at the University of San Diego, California, USA, and the founder and director of Lemur Love, a US-based nonprofit organization conducting research, conservation, and small-scale development in Madagascar. Her research examines the ecology of wild ring-tailed lemurs and the legal and illegal trades of wild-captured lemurs in Madagascar.
Acknowledgments
We thank Madagascar Ministry of the Environment and Forests for permission to study ring-tailed lemurs (no. 155/19/MEDD/SG/DGEF/DGRNE) and the lemur facility for permission to conduct surgery and sample the sick ring-tailed lemurs.
The University of San Diego and Lemur Love, Inc. provided funding for this study.
References
- Blancou J, Rakotoniaina P, Cheneau Y. Types of TB bacteria in humans and animals in Madagascar [in French]. Arch Inst Pasteur Madagascar. 1974;43:31–8.
- Ligthelm LJ, Nicol MP, Hoek KGP, Jacobson R, van Helden PD, Marais BJ, et al. Xpert MTB/RIF for rapid diagnosis of tuberculous lymphadenitis from fine-needle-aspiration biopsy specimens. J Clin Microbiol. 2011;49:3967–70. DOIPubMedGoogle Scholar
- Hunt M, Bradley P, Lapierre SG, Heys S, Thomsit M, Hall MB, et al. Antibiotic resistance prediction for Mycobacterium tuberculosis from genome sequence data with Mykrobe. Wellcome Open Res. 2019;4:191. DOIPubMedGoogle Scholar
- Shitikov E, Kolchenko S, Mokrousov I, Bespyatykh J, Ischenko D, Ilina E, et al. Evolutionary pathway analysis and unified classification of East Asian lineage of Mycobacterium tuberculosis. Sci Rep. 2017;7:9227. DOIPubMedGoogle Scholar
- Rutaihwa LK, Menardo F, Stucki D, Gygli SM, Ley SD, Malla B, et al. Multiple introductions of Mycobacterium tuberculosis lineage 2–Beijing into Africa over centuries. Front Ecol Evol. 2019;7:112. DOIGoogle Scholar
- Stucki D, Brites D, Jeljeli L, Coscolla M, Liu Q, Trauner A, et al. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat Genet. 2016;48:1535–43. DOIPubMedGoogle Scholar
- Ferdinand S, Sola C, Chanteau S, Ramarokoto H, Rasolonavalona T, Rasolofo-Razanamparany V, et al. A study of spoligotyping-defined Mycobacterium tuberculosis clades in relation to the origin of peopling and the demographic history in Madagascar. Infect Genet Evol. 2005;5:340–8. DOIPubMedGoogle Scholar
- LaFleur M, Clarke TA, Reuter KE, Schaefer MS, terHorst C. Illegal trade of wild-captured Lemur catta within Madagascar. Folia Primatol (Basel). 2019;90:199–214. DOIPubMedGoogle Scholar
- Reuter KE, LaFleur M, Clarke TA, Holiniaina Kjeldgaard F, Ramanantenasoa I, Ratolojanahary T, et al. A national survey of household pet lemur ownership in Madagascar. PLoS One. 2019;14:
e0216593 . DOIPubMedGoogle Scholar - Chomel BB, Belotto A, Meslin FX. Wildlife, exotic pets, and emerging zoonoses. Emerg Infect Dis. 2007;13:6–11. DOIPubMedGoogle Scholar
Figure
Cite This ArticleOriginal Publication Date: February 14, 2021
Table of Contents – Volume 27, Number 3—March 2021
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Marni LaFleur, Department of Anthropology, University of San Diego, Saints Tekawitha-Serra Hall, Suite 218C, 5998 Alcala Park, San Diego, CA 92110-2492, USA
Top