Volume 27, Number 3—March 2021
Research Letter
Local Transmission of SARS-CoV-2 Lineage B.1.1.7, Brazil, December 2020
Figure
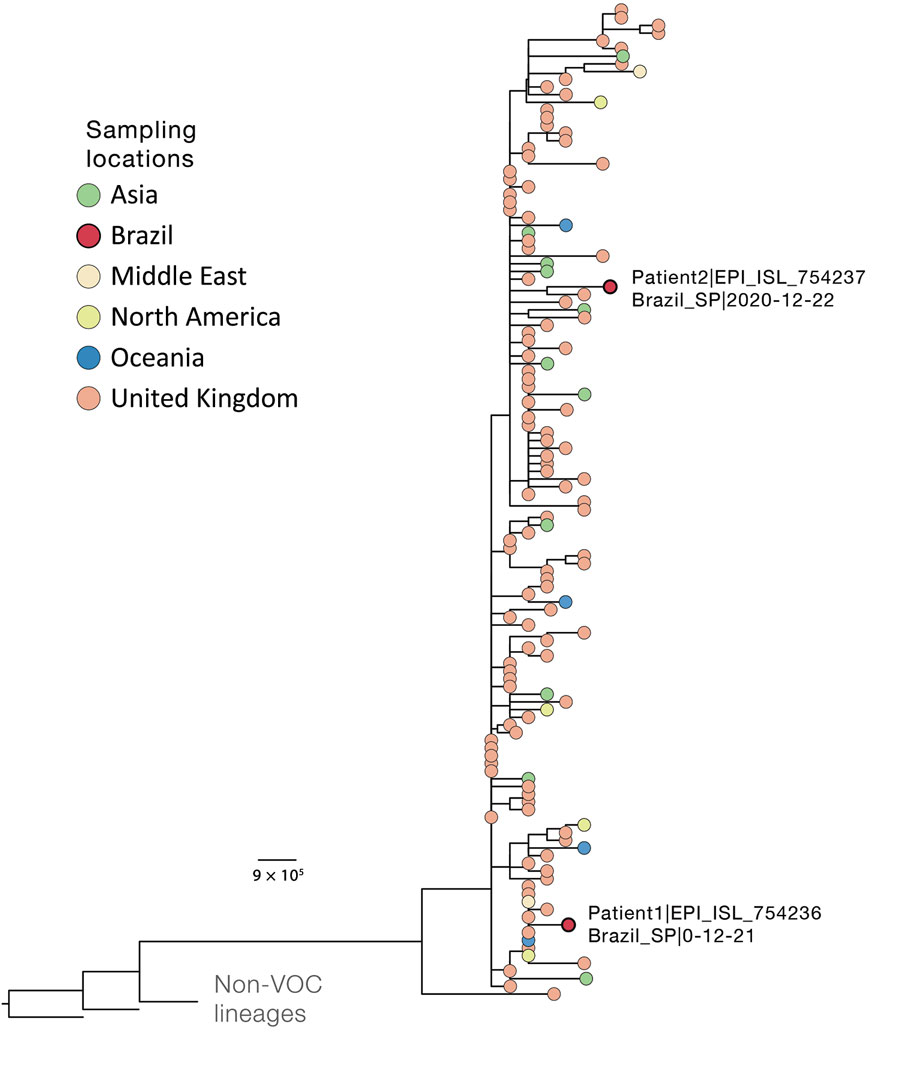
Figure. Phylogenetic context of novel severe acute respiratory syndrome coronavirus 2 B.1.1.7 genomes isolated from 2 patients in Brazil (labeled on figure), December 2020. Downsampling for the phylogenetic analysis of the B.1.1.7 SARS-CoV-2 variant (n = 4,693, December 31, 2020) was performed by selecting 1 sequence per country per day. As outgroups, we included 2 B.1.1 sequences from the United Kingdom that were closely related to the lineage of interest and sequence WH04 from Wuhan, China (GISAID identification no. EPI_ISL_406801; http://www.gisaid.org). Details on multiple alignment and phylogenetic tree reconstruction are described elsewhere (4). Tree file, aligned sequences, and GISAID acknowledgment tables are available at https://github.com/CADDE-CENTRE/VOC-Lineage-Brazil. Scale bar indicates nucleotide substitutions per site. VOC, variant of concern.
References
- Vogels CBF, Watkins AE, Harden CA, Brackney DE, Shafer J, Wang J, et al. SalivaDirect: a simplified and flexible platform to enhance SARS-CoV-2 testing capacity. Med. 2020 Dec 26 [Epub ahead of print].
- Rambaut A, Holmes EC, O’Toole Á, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–7. DOIPubMedGoogle Scholar
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4. DOIPubMedGoogle Scholar
- Candido DS, Claro IM, de Jesus JG, Souza WM, Moreira FRR, Dellicour S, et al.; Brazil-UK Centre for Arbovirus Discovery, Diagnosis, Genomics and Epidemiology (CADDE) Genomic Network. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369:1255–60. DOIPubMedGoogle Scholar
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. DOIPubMedGoogle Scholar
- Shu Y, McCauley J. GISAID: Global Initiative on Sharing All Influenza Data—from vision to reality. Euro Surveill. 2017;30;22:30494.
1These first authors contributed equally to this article.
2These senior authors contributed equally to this article.