Volume 27, Number 5—May 2021
Research
Genetic Evidence and Host Immune Response in Persons Reinfected with SARS-CoV-2, Brazil
Abstract
The dynamics underlying severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfection remain poorly understood. We identified a small cluster of patients in Brazil who experienced 2 episodes of coronavirus disease (COVID-19) in March and late May 2020. In the first episode, patients manifested an enhanced innate response compared with healthy persons, but neutralizing humoral immunity was not fully achieved. The second episode was associated with different SARS-CoV-2 strains, higher viral loads, and clinical symptoms. Our finding that persons with mild COVID-19 may have controlled SARS-CoV-2 replication without developing detectable humoral immunity suggests that reinfection is more frequent than supposed, but this hypothesis is not well documented.
Confirmed cases of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have surpassed 110 million, along with 2.5 million deaths by 2019 coronavirus disease (COVID-19) (1). New waves of the pandemics in different Northern and Southern Hemisphere countries provide evidence that herd immunity might not have been fully achieved and that new variants could escape the response to natural infection (2,3).
Although there is evidence of the generation of B and T memory cells to SARS-CoV-2 proteins after infection (4,5), it has also been documented that neutralizing seroconversion is heterogeneous among the population (6). Even for those who seroconvert, the sustainability of the immune response, as judged by IgG level, might decay after the primary exposure to coronaviruses (7–9). Cases of reinfection by SARS-CoV-2 can be associated with the absence of neutralizing serologic titers, diminishment of immunoglobulin titers after primo-infection, or viral polymorphisms to escape the host SARS-CoV-2 immune response (10–16).
To better understand the dynamics of the immune and virological responses in mild cases of COVID-19 that might predispose patients to reinfection, we continuously followed up with patients for potential exposure to SARS-CoV-2. For 2 patients, reinfection was documented. The National Review Board of Brazil approved the study protocol (Comissão Nacional de Ética em Pesquisa [CONEP] 30650420.4.1001.0008), and informed consent was obtained from all participants or patients’ representatives.
Ethics and Study Population
During March–December 2020, the COVID-19 research task force screened a group of 30 participants weekly, independent of any symptoms, for SARS-CoV-2 detection by RT-PCR in nasopharyngeal swab specimens. If any of these participants exhibited positive results, or members of their households experienced signs or symptoms of COVID-19, they were invited to participate in the study and follow-up. At baseline and follow-up, we collected plasma, serum, and nasopharyngeal swab samples biweekly or at longer intervals if the patient was unavailable (Table). Households were included upon their request to be tested for SARS-CoV-2. Among the participants, 4 exhibited >1 episode of mild self-limiting COVID-19 with positive RT-PCR. For comparison, we included age-matched controls from the same group of participants and city in which the patients lived, Rio de Janeiro, Brazil. Controls were composed of 5 persons negative for SARS-CoV-2 throughout the investigated period.
Measurement of Serum SARS-CoV-2 Antibodies and Plasma Cytokine Levels
For quantitative analysis of SARS-CoV-2 spike protein IgM, IgA, and IgG antibodies, we performed the S-UFRJ test developed at Universidade Federal do Rio de Janeiro (R.G.F. Alvim et al., unpub. data, https://doi.org/10.1101/2020.07.13.20152884) (Appendix).
We collected plasma samples in tubes containing EDTA. We used commercial ELISA kits from R&D Systems (https://www.rndsystems.com) to measure cytokines and chemokine (Appendix).
Molecular Diagnosis
To determine serum titers to block SARS-CoV-2 infection, we performed miniaturized plaque-reduction neutralization test (PRNT) (Appendix). SARS-CoV-2 RNA has been detected in accordance with the US Centers for Disease Control and Prevention (CDC) recommendation (17). We used the standard curve method for virus quantification, using synthetic RNA for gene N (Microbiologics, https://www.microbiologics.com). We compared cycle thresholds (Ct) for the target gene to those obtained with different cell amounts (107–102), for reaction calibration (Appendix).
Genomic Analysis
We extracted total viral RNA from nasopharyngeal swabs using QIAamp Viral RNA (QIAGEN, https://www.qiagen.com), with minor modifications (18) (Appendix). We performed an amplicon-based enrichment strategy using the ATOPlex SARS-CoV-2 Full-Length Genome Panel version 1.0 (MGI Tech Co., https://en.mgi-tech.com; donated by the vendor). Single-stranded circular DNA library pools were converted to DNA nanoballs by rolling circle amplification and submitted to pair-end sequencing (100 nt) on the MGISEQ-2000 platform (recently named DNBSEQ-G400; MGI Tech Co. Ltd.).
We quality-scored, filtered, trimmed, and assembled genomic sequences in contigs through a validated workflow for SARS-CoV-2 (19). Genomes were aligned with MAFFT (20) or ClustalW (21), and phylogenies were constructed with MEGA version 7.0 (22,23), using the Jukes-Cantor model for maximum-likelihood estimates by applying neighbor-joining and BioNJ algorithms (24), or by MrBayes version 3.2.7 (http://nbisweden.github.io/MrBayes) (25,26) with a relaxed clock model with a priori model testing using the gamma rates and invariant sites nucleotide substitution model, selected by jModelTest version 1.6 http://darwin.uvigo.es/software/jmodeltest.htm. We visualized and edited the tree with FigTree version 1.4.2 (http://tree.bio.ed.ac.uk). We determined SARS-CoV-2 clades using the Nextclade software, beta version 0.14 (https://clades.nextstrain.org). To categorize mutations and polymorphisms, we aligned the SARS-CoV-2 reference genome Wuhan-Hu-1 (GISAID EPI ISL no. 402125; https://www.gisaid.org) to our sequences. The original sequences used in this work are publicly available on https://nextstrain.org/ncov: GISAID EPI ISL nos. 636737, 636834–636838. The dataset included in the analysis contained representative sequences of the emerging clades associated with our sequences, 19A and 20B, as well as sequences from the genome 20A as a negative control (Appendix Table 1).
Among the households of the COVID-19 research task force, a 54-year-old man (patient A) requested an RT-PCR test for SARS-CoV-2 on March 23 because of a recurrent headache on the prior 2 days. He also had previous contact with a symptomatic co-worker returning from travel who refused to be tested. Patient A had a detectable viral load (Ct 27.41) of ≈105 copies/mL in nasopharyngeal swab samples (Table). Although patient B, a 57-year-old woman with a previous history of discoid lupus erythematosus, was in self-isolation, she was tested because of close contact with patient A. She tested positive for COVID-19 on March 24; her nasopharyngeal swab sample Ct was ≈36.31 (≈103 copies/mL) (Table). Two days afterward, she experienced diarrhea (Table).
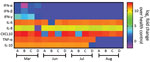
Patient B shares a household with patients C and D, a married couple, both 34 years old. Patients C and D were not in social isolation because of their work duties. Although patient C was asymptomatic, he displayed a Ct of 35.71 (103 copies/mL) on March 25 (Table). Patient D was negative by molecular testing on March 26, but 1 week later, she had a detectable viral load (Ct 36.01, 103 copies/mL) and reported diarrhea in the following days (Table). On March 27, all 4 patients experienced an increase of inflammatory mediators (interleukin [IL] 6, IL-8, and tumor necrosis factor α) and regulatory (IL-10) and chemotactic (C-X-C motif chemokine ligand 10) and antiviral (interferon γ) signals, relative to healthy SARS-CoV-2–negative controls (Figure 1). Although cytokine response was consistent with the resolution of the infection, the anti–SARS-CoV-2 neutralizing humoral response was not detected in late March 2020 (Table; Appendix Figure 2).
For patients B and C, we were able to obtain a full-length SARS-CoV-2 genome (Table). Complete genome sequencing, with Phred quality score >30, composed of 140,000–20,000,000 reads and 100-fold to 10,000-fold coverage, argues against a false-positive RT-PCR result (Appendix Table 2, first column). For patients A and D, the samples were insufficient for sequencing. In March 2020, patients B was infected with emerging SARS-CoV-2 clade 19A and patient C with SARS-CoV-2 clade 20B, (Table; Figure 2; Appendix Figure 3). The detection of the 2 distinct lineages indicates that patients B and C were infected independently and did not transmit the virus to each other (Table; Figure 2; Appendix Figure 3). These distinct lineages were co-circulating in Brazil in March 2020 when multiple introductions of the SARS-CoV-2 occurred (27). Emerging clade 19A is associated with imported cases in Brazil, because of its proximity to the Wuhan-01 sequence (Figure 2; Appendix Figure 3). Indeed, detection of clade 19A in the sample from Patient B is consistent with household transmission from patient A, and his contact with the symptomatic traveler. Patient C, a police officer, was frequently exposed to various probable sources of contamination; he was infected with an emerging clade 20B virus, the most prevalent variant in Brazil, during December 2020 (Figure 2, Appendix Figure 3). All patients recovered from a mild COVID-19 episode and were retested in the first half of April, when they had negative RT-PCR results.
In the last week of May 2020, when COVID-19 cases in Rio de Janeiro were at the peak of the first wave of the pandemic (28), these 4 patients reported more signs and symptoms of SARS-CoV-2 infection than in March (Table). During the second episode, they experienced fever and cough, along with fatigue, headache, body ache, anosmia, and ageusia. Real-time RT-PCR revealed higher viral loads in the nasopharyngeal swab samples than at the time of the first infection: Ct of 21.76 (≈107 copies/mL) for patient A, 21.84 (≈107 copies/mL) for patient B, 26.38 (≈105 copies/mL) for patient C, and 16.87 (≈109 copies/mL) for patient D (Table).
On June 3, a week after the second episode, we detected SARS-CoV-2 immunoglobulins in patients A and B, but they had low to no neutralizing activity (Table; Appendix Figure 2). These serologic samples from June indicate that the first episode of COVID-19 was not followed by a sustained neutralizing humoral response, as judged by 90% PRNT (PRNT90) titers (Table). Because signals of a humoral effector memory were inconsistent after the first episode of COVID-19 (Table), we could speculate that the enhanced production of interferons and proinflammatory mediators led to resolution of the primo-infection (Figure 1). During the second episode of COVID-19, most of the cytokine levels were still higher than in healthy volunteers (Figure 1).
On July 9, forty days after the episode of reinfection, all patients had detectable immunoglobulin levels and their lowest PRNT90 results (Table; Appendix Figure 2), declining thereafter by August 10 (Table; Appendix Figure 2). In July, patients’ tests continuously showed upregulated pro-inflammatory markers (Figure 1), which are consistent with an enhanced response to a second SARS-CoV-2 exposure. In August, the markers of inflammation and regulatory responses, tumor necrosis factor α and IL-10, decreased compared with levels from previous months (Figure 1).
In the second episode, we fully sequenced the SARS-CoV-2 genome from all patients (Table; Figure 2; Appendix Table 2, Figure 3). SARS-CoV-2 sequences from the reinfection clustered together, suggesting a household transmission for patients A–D (Figure 2; Appendix Figure 3). The emerging genotype 20B, which was the main variant circulating in Brazil since May 2020, was detected in all samples from the second episode (Figure 2; Table; Appendix Figure 3). For patient B, the first episode was associated with the emerging clade 19A and the second with 20B (Figure 2; Appendix Figure 3). Two episodes provoked by genetically distinct lineages support the possibility of reinfection.
Although both episodes in patient C were associated with clade 20B, they clustered apart on the phylogeny with significant statistical support: by 86% of bootstrap using maximum likelihood (Figure 2) and by Bayesian inference (Appendix Figure 3). Genetic markers in the SARS-CoV-2 genome were different in the patient’s 2 episodes of COVID-19 (Appendix Table 2). The genomes diverge at the genes encoding the nonstructural protein (NSP) 3, 3C-like proteinase, and exonuclease (Appendix Table 2). In addition to the genetic variations, poor development of anti–SARS-CoV-2 serology between the 2 episodes of infection points suggests a reinfection scenario.
Seasonal human coronaviruses may cause reinfection, as documented for the past 35 years (8,29). Of note, in veterinary medicine, domestic mammals also have coronavirus reinfection (30). Adaptive, memory-generating immunity to coronaviruses is heterogeneously sustainable in mammals, and some events of infection are controlled at the level of the innate immunity (31–33). We fully documented reinfection in 2 genetically unrelated persons in Rio de Janeiro, Brazil, describing patients who sought care twice in a 2-month interval who received clinical and laboratory diagnosis of COVID-19. Virus polymorphisms from the primary and second episodes and negative RT-PCR between the events strengthen the argument toward reinfection. Neutralizing anti–SARS-CoV-2 titers were not detected during the first episode, nor at the baseline of the second episode, suggesting that patients were still vulnerable after the primary episode.
SARS-CoV-2 reinfection has been associated with new variants that overcome the immune response to natural infection, short-lasting humoral response, and a limited or absent neutralizing immunity after the primo-infection (10–13). The patients in Brazil described in this study are similar to cases in the United States and Ecuador (10,13), in which reinfection was associated with more symptoms. Antibody-dependent enhancement or exposure to higher amounts of the virus could be the reason for the change from asymptomatic or oligosymptomatic to syndromic. In our study, primary and second infections were caused by a strain carrying the D614G mutation in the spike protein, which has been associated with higher replication efficiency (34). We did not detect other contemporaneous changes in the spike protein, such as 69/70 deletion, K417N, E484K, N501Y, P681H, or the 17 unique mutations of the P1 variant, which precluded association with more virulent strains (35). Beyond the spike protein, we detected the V125F change in the NSP14 protein; V125F is a nonconservative mutation that might increase the volume in the loop between β-sheets number 5 and 6, which could affect its methyltransferase activity (36). The V125F mutation is unlikely to increase virulence in a second episode. On the other hand, changes in NSP6 protein (37) and open reading frame 6 mRNA (S. Sarif Hassan et al., unpub. data, https://doi.org/10.1101/2020.11.06.372227) might result in viral evasion from innate immunity.
The primary infections of patients B and C were associated with emerging clades 19A and 20B, indicating that the 2 cohabitants were infected independently. Indeed, while 1 patient was in social isolation, the others were working outside in the community. The cocirculation of these clades of SARS-CoV-2 is consistent with the COVID-19 databases in GISAID and the multiple introductions of the new coronavirus in Brazil (27). In the following months, emerging clade 20B was identified as the most prevalent genotype, representing 60% of the deposited genomes on GISAID. The detection of clade 20B on the second episode of COVID-19, by the end of May, is associated with the peak of the pandemic in Rio de Janeiro, Brazil (28).
Distinct clades of SARS-CoV-2 were found in the primary and secondary respiratory samples from patient B, supporting the notion of reinfection. For patient C, both the first and second detections of SARS-CoV-2 were associated with clade 20B. Although viral persistence could be imagined in this scenario, SARS-CoV-2 genomic sequences from the first and second episodes do not cluster together in the same branch, as they did for an immunocompromised patient that shed SARS-CoV-2 for 150 days (38). Thus, phylogeny does not support the interpretation of persistence, by different methods. By branching apart, SARS-CoV-2 genomes associated with patient C strengthen the chances of a relevant degree of variation (39), indicating the direction of reinfection. In the documented case of SARS-CoV-2 and human coronavirus NL63 reinfection, different episodes were genetically associated with similar viral clades or strains (40). Whereas the detection of 2 episodes of SARS-CoV-2 infection from patient C was separated by >60 days, prolonged virus shedding in the nasopharyngeal swab specimens from mild cases lasted for 22–46 days (41), which is further evidence against persistence.
Results of SARS-CoV-2 reinfection affirm that immune rechallenge may be necessary to achieve humoral protection and underscore that sustainability of the immune response may be heterogeneous. We documented that these patients with mild COVID-19 displayed an innate immune response composed of pro-inflammatory and regulatory signals. Although cytokine storm has been associated with severe COVID-19 (42), we interpret that in the case of our patients, the innate immune response might have led to infection resolution (43). Another possibility, not explored in detail here, is that cellular-mediated immunity could have contributed to the mild clinical outcome (2,4,44). The natural history of mild COVID-19 described for these patients might also be representative of many persons exposed to the first wave of the pandemic, leading to the hypothesis that they would also be susceptible to other episodes of SARS-CoV-2 infections, even without the challenge being imposed by new variants.
We determined, on the basis of 6 years of surveillance and follow-up of human coronavirus reinfections, that initial exposure was insufficient to elicit a protective immune response, imposing limited pressure on selection on new seasonal coronavirus variants (40). Similarly, our data on a small cluster of patients recapitulate this natural history of reinfection, which may also occur for SARS-CoV-2.
Dr. Fintelman-Rodrigues is based at the Laboratory of Immunopharmacology, Instituto Oswaldo Cruz (IOC), Fundação Oswaldo Cruz (Fiocruz), Rio de Janeiro, Brazil.
Acknowledgments
We thank Carmen Beatriz Wagner and Giacoia Gripp for assessments related to the Biosafety Level 3 facility and Marco Alberto Medeiros for assessments related to the sequencing platform. We thank Gonzalo Bello, Dumith Chequer Bou-Habib, Willian Provance, and Fabiano Thompson for insightful discussions. We greatly appreciated the MGI, a partner in the implementation of next-generation sequencing through collaborations with Oswaldo Cruz Foundation, especially for challenging samples of COVID-19.
Author contributions: F.A.B. and P.T.B. conducted clinical surveillance. N.F.R. and F.A.B. enrolled patients in the study. N.F.R., C.Q.S., D.R., I.G.A., V.C.S. performed immunologic assessments. N.F.R., A.P.D.S., M.C.S., F.B.S., M.A.F., J.G., H.J., and H.T. performed sequencing. A.P.D.S., M.C.S., and D.A.T were responsible for bioinformatics. F.A.B., P.T.B., C.M.M., T.M.L.S. handled study coordination. N.F.R., P.T.B., A.MV., and T.M.L.S. prepared and revised the manuscript. All authors revised and approved the manuscript.
References
- World Health Organization. Coronavirus disease (COVID-19) dashboard. 2020 [cited 2020 Nov 12]. https://covid19.who.int/
- Rodda LB, Netland J, Shehata L, Pruner KB, Morawski PA, Thouvenel CD, et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 2021;184:169–183.e17. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Cases, data, and surveillance. 2020 [cited 2021 Feb 12]. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html
- Hartley GE, Edwards ESJ, Aui PM, Varese N, Stojanovic S, McMahon J, et al. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci Immunol. 2020;5:
eabf8891 . DOIPubMedGoogle Scholar - Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–62. DOIPubMedGoogle Scholar
- Ripperger TJ, Uhrlaub JL, Watanabe M, Wong R, Castaneda Y, Pizzato HA, et al. Detection, prevalence, and duration of humoral responses to SARS-CoV-2 under conditions of limited population exposure. Immunity. 2020;53:925–33. DOIPubMedGoogle Scholar
- Hueston L, Kok J, Guibone A, McDonald D, Hone G, Goodwin J, et al. The antibody response to SARS-CoV-2 infection. Open Forum Infect Dis. 2020;7:a387. DOIPubMedGoogle Scholar
- Edridge AWD, Kaczorowska J, Hoste ACR, Bakker M, Klein M, Loens K, et al. Seasonal coronavirus protective immunity is short-lasting. Nat Med. 2020;26:1691–3. DOIPubMedGoogle Scholar
- Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:
eabf4063 . DOIPubMedGoogle Scholar - Tillett RL, Sevinsky JR, Hartley PD, Kerwin H, Crawford N, Gorzalski A, et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2021;21:52–8. DOIPubMedGoogle Scholar
- To KK-W, Hung IF-N, Ip JD, Chu AW-H, Chan W-M, Tam AR, et al. Coronavirus disease 2019 (COVID-19) reinfection by a phylogenetically distinct severe acute respiratory syndrome coronavirus 2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2020;•••:
ciaa1275 ; Epub ahead of print. DOIGoogle Scholar - Van Elslande J, Vermeersch P, Vandervoort K, Wawina-Bokalanga T, Vanmechelen B, Wollants E, et al. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin Infect Dis. 2020;•••:
ciaa1330 ; Epub ahead of print. DOIPubMedGoogle Scholar - Prado-Vivar B, Becerra-Wong M, Guadalupe JJ, Marquez S, Gutierrez B, Rojas-Silva P, et al. COVID-19 reinfection by a phylogenetically distinct SARS-CoV-2 variant, first confirmed event in South America. SSRN. 2020 September 9 [cited 2021 Mar 19]. DOIGoogle Scholar
- Mulder M, van der Vegt DSJM, Oude Munnink BB, GeurtsvanKessel CH, van de Bovenkamp J, Sikkema RS, et al. Reinfection of severe acute respiratory syndrome coronavirus 2 in an immunocompromised patient: a case report. Clin Infect Dis. 2020;•••:
ciaa1538 ; Epub ahead of print. DOIGoogle Scholar - Selhorst P, Van Ierssel S, Michiels J, Mariën J, Bartholomeeusen K, Dirinck E, et al. Symptomatic SARS-CoV-2 reinfection of a health care worker in a Belgian nosocomial outbreak despite primary neutralizing antibody response. Clin Infect Dis. 2020;•••:
ciaa1850 ; Epub ahead of print. DOIPubMedGoogle Scholar - Larson D, Brodniak SL, Voegtly LJ, Cer RZ, Glang LA, Malagon FJ, et al. A case of early reinfection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis. 2020;•••:
ciaa1436 ; Epub ahead of print. DOIGoogle Scholar - Centers for Disease Control and Prevention. Research use only 2019-novel coronavirus (2019-nCoV) real-time RT-PCR primers and probes. 2020 [cited 2020 Nov 11]. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html
- Metsky HC, Matranga CB, Wohl S, Schaffner SF, Freije CA, Winnicki SM, et al. Zika virus evolution and spread in the Americas. Nature. 2017;546:411–5. DOIPubMedGoogle Scholar
- Cleemput S, Dumon W, Fonseca V, Abdool Karim W, Giovanetti M, Alcantara LC, et al. Genome Detective Coronavirus Typing Tool for rapid identification and characterization of novel coronavirus genomes. Bioinformatics. 2020;36:3552–5. DOIPubMedGoogle Scholar
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–8. DOIPubMedGoogle Scholar
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. DOIPubMedGoogle Scholar
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. DOIPubMedGoogle Scholar
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91. DOIPubMedGoogle Scholar
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Mammalian protein metabolism. Vol. III. Munro HN, editor. New York: Academic Press; 1969. p. 21–132.
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5. DOIPubMedGoogle Scholar
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. DOIPubMedGoogle Scholar
- Candido DS, Claro IM, de Jesus JG, Souza WM, Moreira FRR, Dellicour S, et al.; Brazil-UK Centre for Arbovirus Discovery, Diagnosis, Genomics and Epidemiology (CADDE) Genomic Network. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;369:1255–60. DOIPubMedGoogle Scholar
- Secretaria de Saúde do Estado do Rio de Janeiro. Covid-19 monitoring panel in the Rio de Janeiro State [in Portuguese]. 2020 [cited 2020 Nov 24]. http://painel.saude.rj.gov.br/monitoramento/covid19.html#
- Kiyuka PK, Agoti CN, Munywoki PK, Njeru R, Bett A, Otieno JR, et al. Human coronavirus NL63 molecular epidemiology and evolutionary patterns in rural coastal Kenya. J Infect Dis. 2018;217:1728–39 https://academic.oup.com/jid/article/217/11/1728/4948258. DOIPubMedGoogle Scholar
- Decaro N, Martella V, Saif LJ, Buonavoglia C. COVID-19 from veterinary medicine and one health perspectives: What animal coronaviruses have taught us. Res Vet Sci. 2020;131:21–3. DOIPubMedGoogle Scholar
- Neeland MR, Bannister S, Clifford V, Dohle K, Mulholland K, Sutton P, et al. Innate cell profiles during the acute and convalescent phase of SARS-CoV-2 infection in children. Nat Commun. 2021;12:1084. DOIPubMedGoogle Scholar
- Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–74. DOIPubMedGoogle Scholar
- Sallenave J-M, Guillot L. Innate immune signaling and proteolytic pathways in the resolution or exacerbation of SARS-CoV-2 in COVID-19: key therapeutic targets? Front Immunol. 2020;11:1229. DOIPubMedGoogle Scholar
- Groves DC, Rowland-Jones SL, Angyal A. The D614G mutations in the SARS-CoV-2 spike protein: Implications for viral infectivity, disease severity and vaccine design. Biochem Biophys Res Commun. 2021;538:104–7. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Emerging SARS-CoV-2 variants. 2020 [cited 2021 Feb 12]. https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html
- Krafcikova P, Silhan J, Nencka R, Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat Commun. 2020;11:3717. DOIPubMedGoogle Scholar
- Cottam EM, Whelband MC, Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10:1426–41. DOIPubMedGoogle Scholar
- Choi B, Choudhary MC, Regan J, Sparks JA, Padera RF, Qiu X, et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med. 2020;383:2291–3. DOIPubMedGoogle Scholar
- Koyama T, Platt D, Parida L. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ. 2020;98:495–504. DOIPubMedGoogle Scholar
- Kiyuka PK, Agoti CN, Munywoki PK, Njeru R, Bett A, Otieno JR, et al. Human coronavirus NL63 molecular epidemiology and evolutionary patterns in rural coastal Kenya. J Infect Dis. 2018;217:1728–39. DOIPubMedGoogle Scholar
- Sun J, Xiao J, Sun R, Tang X, Liang C, Lin H, et al. Prolonged persistence of SARS-CoV-2 RNA in body fluids. Emerg Infect Dis. 2020;26:1834–8. DOIPubMedGoogle Scholar
- de la Rica R, Borges M, Gonzalez-Freire M. COVID-19: in the eye of the cytokine storm. Front Immunol. 2020;11:
558898 . DOIPubMedGoogle Scholar - Arunachalam PS, Wimmers F, Mok CKP, Perera RAPM, Scott M, Hagan T, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–20. DOIPubMedGoogle Scholar
- Sekine T, Perez-Potti A, Rivera-Ballesteros O, Strålin K, Gorin J-B, Olsson A, et al.; Karolinska COVID-19 Study Group. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: April 01, 2021
Table of Contents – Volume 27, Number 5—May 2021
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Thiago Moreno Lopes e Souza, Fundação Oswaldo Cruz (Fiocruz), Centro de Desenvolvimento Tecnológico em Saúde (CDTS), Instituto Oswaldo Cruz (IOC), Pavilhão Osório de Almeida, sala 16, Av. Brasil 4365, Manguinhos, Rio de Janeiro, RJ, CEP 21060340, Brazil
Top