Volume 27, Number 7—July 2021
Research Letter
Outbreak of Rabbit Hemorrhagic Disease Virus 2 Infections, Ghana
Cite This Article
Citation for Media
Abstract
In September 2019, high mortality in commercial rabbits was reported in the Greater Accra Region of Ghana. Rabbit hemorrhagic disease virus 2 phylogenetically related to isolates from 2015–2017 outbreaks in the Netherlands was confirmed as the causative agent. The virus has not yet been detected in native rabbits in Ghana.
Rabbit hemorrhagic disease (RHD) is an acute, fatal, highly contagious viral hepatitis in European rabbits (Oryctolagus cuniculus) (1). It causes severe economic losses in the rabbit meat and fur industries and can have a substantial negative ecological impact on wild rabbit populations and their predators. The causative agent, RHD virus (RHDV; family Caliciviridae, genus Lagovirus) has a single-stranded positive-sense RNA genome ≈7.4 kb in length. Pathogenic RHDV strains exist in 2 main genotypes, GI.1 and GI.2 (RHDV2). RHDV GI.1 has several variants: GI.1a (proposed as G6/RHDVa), GI.1b (G1), GI.1c (G2), and GI.1d (G3–G5) (2). It is considered enzootic in domestic and wild European rabbits in Asia and Europe; sporadic outbreaks occur in the Americas, Middle East, and Africa (1). RHDV2 GI.2, a variant first reported in France in 2010 (3), differs antigenically from RHDV GI.1 and is therefore considered a distinct serotype. RHDV2, which has replaced RHDV GI.1 in many countries in Europe, infects rabbits of all ages and crosses the species barrier to affect non-European rabbit species (4).
Rabbit production, because of its low costs, has been promoted to reduce poverty in Africa but has been threatened by RHD outbreaks since the late 1980s. In 2015, RHDV2 was detected on Tenerife in the Canary Islands (5). In April 2017, an outbreak was reported in northern Morocco (6) caused by a recombinant GI.1b/GI.1b/GI.2 RHDV2 strain closely related to isolates identified in Portugal in 2014. During 2015–2018, RHDV2 strains similar to those circulating in France, Portugal, Spain, and Finland were isolated in field rabbits in Tunisia (7). Recently, rabbit farms in West Africa have been severely affected by RHD. Using competitive ELISA, researchers in Benin in July 2015 confirmed an RHD outbreak in the coastal city Cotonou (8). In February 2016, several RHD outbreaks detected by using real-time reverse transcription PCR (rRT-PCR) in Korhogo in northern Côte d’Ivoire, were resolved but the virus and rabbit species were not well characterized (8). On June 3, 2020, Senegal reported its first RHDV2 outbreak, which killed many farmed Flemish giant, checkered giant, and Fauve de Bourgogne rabbits (8). Nigeria’s first reported outbreak started in September 2020 at rabbit farms in Kwara and Oya, southwestern states bordering Benin (8).
A September 2019 RHDV2 outbreak sickened 11,350 rabbits and killed ≈6,000 in commercial rabbit farms in Dansoman, Korle-Bu, Mamprobi, and Banana Inn in the Ablekuma Central District, Greater Accra Region, Ghana. Clinical signs included dullness, anorexia, convulsion, dyspnea, bloody nasal discharge, mucus in feces, and paralysis. Postmortem examinations revealed multifocal petechial hemorrhages in multiple organs, congested and edematous lungs, pulmonary edema with tracheal foam, splenomegaly, and pale livers with areas of necrosis. Tissue samples from affected animals submitted to the Accra Veterinary Laboratory tested positive for RHDV2. The disease quickly spread to other areas of Ghana, including the Ashanti Central Eastern, Bono, and Ahafo regions. We sent 35 rRT-PCR–positive liver, lung, and spleen samples from different regions of Ghana (Appendix Figure 1) to the National Centre for Foreign Animal Disease in Winnipeg, Manitoba, Canada, where rRT-PCR testing confirmed RHDV2 (9).
We extracted nucleic acid from 1 of the samples (RHDV2 Ghana-15 2019; Dansoman, Ghana) with a cycle threshold of 12.73 and subjected it to whole-genome sequencing on an Illumina MiSeq instrument (https://www.illumina.com) after targeted cDNA conversion as described elsewhere (10) with modifications. The whole genome sequence (GenBank accession no. MW118115) closely resembled (98.84%) that of RHDV2 isolate RHDV-NL2016 (accession no. MN061492) from the 2015–2017 outbreak in the Netherlands (Appendix Figure 2). We later subjected 14 additional independent samples from different locations in the greater Accra region to whole-genome sequencing (accession no. MW789232–245) and found they were ≥99.73% identical at the nucleotide level to RHDV2 Ghana-15 2019, suggesting that this outbreak was caused by a single incursion.
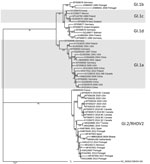
The viral protein 60 gene of RHDV2 Ghana-15 2019 showed 98.73% identity to the same gene in the isolate from the Netherlands (accession no. MN061492), 96.32% to the isolate (accession no. MF407653) from Spain, and only 94.70% to the isolate from Tunisia (accession no. MK629981; Figure). No sequence information was available from the previous RHDV2 outbreaks in West Africa, and therefore it is not possible to comment on the relatedness of the recent RHDV2 outbreaks in this region. Introduction of RHDV2 at the port cities in West Africa suggests that the virus likely entered each country through imported rabbits or rabbit byproducts and was not spread by rabbits moving across land borders. The outbreak in Ghana continues, and the Ghana Veterinary Authority is considering vaccination to protect local rabbit populations. So far, no RHD cases have been reported in native rabbits in Ghana, but the lack of infections could be simply because native rabbits have not yet been exposed to the virus, which could change in the future.
Dr. Ambagala is the section head and the World Organization for Animal Health–designated expert on classical swine fever at the Canadian Food Inspection Agency–National Centre for Foreign Animal Disease, in Winnipeg. Dr. Ambagala’s research focuses on the detection, prevention, and control of imported and emerging viral diseases of farm animals.
Acknowledgment
This study was supported by the Canadian Food Inspection Agency and the Ministry of Food and Agriculture of Ghana. The authors thanks Dr. Paul Polku (Accra Metropolitan Veterinary Officer), Dr. Fenteng Danso (Epidemiology Unit) and Dr. Patrick Amponsah (Kumasi Veterinary Laboratory) at the Ministry of Food and Agriculture of Ghana for their help with sample collection.
References
- Abrantes J, van der Loo W, Le Pendu J, Esteves PJ. Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV): a review. Vet Res (Faisalabad). 2012;43:12. DOIPubMedGoogle Scholar
- Le Pendu J, Abrantes J, Bertagnoli S, Guitton JS, Le Gall-Reculé G, Lopes AM, et al. Proposal for a unified classification system and nomenclature of lagoviruses. J Gen Virol. 2017;98:1658–66. DOIPubMedGoogle Scholar
- Le Gall-Reculé G, Zwingelstein F, Boucher S, Le Normand B, Plassiart G, Portejoie Y, et al. Detection of a new variant of rabbit haemorrhagic disease virus in France. Vet Rec. 2011;168:137–8. DOIPubMedGoogle Scholar
- Rouco C, Aguayo-Adán JA, Santoro S, Abrantes J, Delibes-Mateos M. Worldwide rapid spread of the novel rabbit haemorrhagic disease virus (GI.2/RHDV2/b). Transbound Emerg Dis. 2019;66:1762–4. DOIPubMedGoogle Scholar
- Martin-Alonso A, Martin-Carrillo N, Garcia-Livia K, Valladares B, Foronda P. Emerging rabbit haemorrhagic disease virus 2 (RHDV2) at the gates of the African continent. Infect Genet Evol. 2016;44:46–50. DOIPubMedGoogle Scholar
- Lopes AM, Rouco C, Esteves PJ, Abrantes J. GI.1b/GI.1b/GI.2 recombinant rabbit hemorrhagic disease virus 2 (Lagovirus europaeus/GI.2) in Morocco, Africa. Arch Virol. 2019;164:279–83. DOIPubMedGoogle Scholar
- Rahali N, Sghaier S, Kbaier H, Zanati A, Bahloul C. Genetic characterization and phylogenetic analysis of rabbit hemorrhagic disease virus isolated in Tunisia from 2015 to 2018. Arch Virol. 2019;164:2327–32. DOIPubMedGoogle Scholar
- World Organisation for Animal Health. World Animal Health Information System [cited 2021 Jan 3]. https://wahis.oie.int
- Duarte MD, Carvalho CL, Barros SC, Henriques AM, Ramos F, Fagulha T, et al. A real time Taqman RT-PCR for the detection of rabbit hemorrhagic disease virus 2 (RHDV2). J Virol Methods. 2015;219:90–5. DOIPubMedGoogle Scholar
- Logan G, Freimanis GL, King DJ, Valdazo-González B, Bachanek-Bankowska K, Sanderson ND, et al. A universal protocol to generate consensus level genome sequences for foot-and-mouth disease virus and other positive-sense polyadenylated RNA viruses using the Illumina MiSeq. BMC Genomics. 2014;15:828. DOIPubMedGoogle Scholar
Figure
Cite This ArticleOriginal Publication Date: June 12, 2021
Table of Contents – Volume 27, Number 7—July 2021
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Aruna Ambagala, CFIA-National Centre for Foreign Animal Disease, 1015 Arlington St, Winnipeg, MB R3E 3M4, Canada
Top