Volume 27, Number 8—August 2021
Research Letter
Linezolid- and Multidrug-Resistant Enterococci in Raw Commercial Dog Food, Europe, 2019–2020
Abstract
We describe enterococci in raw-frozen dog food commercialized in Europe as a source of genes encoding resistance to the antibiotic drug linezolid and of strains and plasmids enriched in antibiotic-resistance and virulence genes in hospitalized patients. Whole-genome sequencing was fundamental to linking isolates from dog food to human cases across Europe.
Raw meat–based diets are increasingly popular for feeding dogs, but the extent of antimicrobial-resistant bacteria in raw dog food is rarely addressed globally (1). The Centers for Disease Control and Prevention does not recommend feeding raw diets to pets because of frequent contamination with Salmonella and Listeria (https://www.cdc.gov/healthypets/publications/pet-food-safety.html), but awareness about this issue is not as evident in Europe. Eating raw meat has been considered a risk factor for carriage of clinically relevant ampicillin-resistant (AmpR) Enterococcus faecium and optrA-positive linezolid-resistant E. faecalis in dogs (2,3), but data for commercial pet food are not available. We evaluated multidrug-resistant (MDR) Enterococcus in raw-frozen dog food commercialized in countries in Europe; we focused on transferable linezolid resistance (LinR) genes because linezolid is a last-resort drug to treat gram-positive infections (4).
We purchased 14 raw-frozen dog food samples from the 2 commercially available brands in Portugal in specialized stores (September 2019–January 2020). Brand A (produced in Europe) is available in specialized stores, brand B (produced in the United Kingdom) in specialized stores and online; both are commercialized across different countries in Europe. We enriched samples (25 g) in buffered peptone water (1:10), then in brain–heart infusion broth with or without different antibiotic drugs (ampicillin [16 μg/mL], vancomycin [6 μg/mL], chloramphenicol [16 μg/mL]), and plated them onto Slanetz-Bartley agar with and without the same drug concentrations. We identified isolates with different morphologies per plate by PCR. We performed antibiotic susceptibility testing by disk diffusion using European Committee on Antimicrobial Susceptibility Testing (EUCAST) (5) or Clinical and Laboratory Standards Institute (6) guidelines. We used broth microdilution for linezolid and Etest for ampicillin. We searched acquired LinR genes (optrA/poxtA/cfrA-E) and typed representative isolates by multilocus sequence typing (n = 20; https://www.pubmlst.org) and whole-genome sequencing (LinR E. faecalis [n = 6] and AmpR/LinR E. faecium [n = 5]) using the Hi Seq 2500 Sequencing System (Illumina, https://www.illumina.com). We deposited assemblies (SPAdes version 3.11.1; https://cab.spbu.ru/software/spades) in GenBank (Bioproject PRJNA663240) and characterized them using in silico tools (http://www.genomicepidemiology.org) and in-house databases (7).
All samples carried enterococci resistant to erythromycin, streptomycin, chloramphenicol, and tetracycline; 93% resistant to ampicillin, ciprofloxacin, and quinupristin/dalfopristin; 79% resistant to gentamicin; and 50% resistant to linezolid. We detected acquired LinR genes among 20 MDR isolates from 64% of samples from both brands and with different types of ingredients (Table): optrA (4 E. faecalis, 1 E. faecium), poxtA (2 E. faecium), optrA+poxtA (8 E. faecalis, 3 E. faecium) or optrA+cfrD (2 E. faecalis). Of those, 15 expressed LinR (MIC = 8 mg/L), whereas 5 were susceptible (MIC = 4 mg/L) (Table).
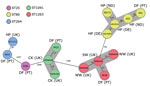
The E. faecium isolates (n = 39) were mostly MDR (70%), expressing resistance to tetracycline (85%), quinupristin/dalfopristin (72%), erythromycin (64%), ciprofloxacin (59%), streptomycin (57%), ampicillin (56%), gentamicin (23%), chloramphenicol (21%), or linezolid (10%). We compared selected dog food AmpR E. faecium genomes with 7,660 available GenBank E. faecium genomes by complex types (CTs) through core-genome multilocus sequence typing (Ridom SeqSphere+ version 7.2, https://www.ridom.de/seqsphere). Those data (Figure) and data from single-nucleotide polymorphisms (Appendix Figure 1) showed different clusters grouping related isolates obtained from dog food and hospitalized patients (sequence type [ST] 80/CT106; ST264/CT374) or from pet food and livestock or wastewaters (ST1091/CT284; ST1263/CT3399) in different countries. Dog food E. faecium was enriched in acquired antibiotic-resistant and virulence genes as strains from different sources (Appendix Figure 1). ST80 E. faecium from brand A was phylogenetically related to other strains from Germany and Netherlands; ST1091 and ST1263 from brand B were phylogenetically related to UK strains (Figure). By filter-mating (8), we found that 3 (ST25, ST80, ST1263) of 5 AmpR E. faecium isolates transferred a chromosomal genetic platform containing pbp5 to GE1 E. faecium strain (Table). Following our previous description of a large transferable pbp5-containing platform in a clinical isolate (8), we partly identified highly similar genetic platforms carrying different adaptive features including virulence genes (e.g., sgrA) in ST80 and ST1263 dog food AmpR E. faecium (Appendix Figure 2). ST1263 E. faecium was able to transfer poxtA by conjugation (Table).
The E. faecalis isolates (n = 52) recovered were mostly MDR (75%), resistant to chloramphenicol (83%), tetracycline (79%), erythromycin (75%), streptomycin (63%), gentamicin (31%), linezolid (21%), or ciprofloxacin (10%). ST40, ST674, ST1008, and ST1009 sequences corresponded to novel complex types carrying antimicrobial resistance (aac(6')-aph(2″)/ant(6)-Ia/aph3″-III/erm(B)/tet(M),tet(L),dfr(G)) and virulence (ace/gelE/elrA) genes linked to clinically relevant MDR lineages (Table) (7,9). ST674 E. faecalis carried optrA on a pheromone-responsive plasmid (pAPT110) identical to others from non–clonally related E. faecalis in hospitalized patients in Spain and China (Appendix Figure 3). Similarly to pAPT110 in this study transferring optrA in high rates (Table), pEF10748 (China) is an optrA highly transferable plasmid with a complete sex-pheromone response module (10).
In conclusion, the diversity and rate of E. faecium and E. faecalis with linezolid-resistance genes (optrA/poxtA/cfrD) we identified were unexpectedly high. Our data suggest that raw dog food could be a sentinel of emerging antimicrobial resistance traits because this type of food may accumulate raw ingredients of different origins, namely from animals associated with intensive farming, adding a new concern to the global health burden of antimicrobial resistance.
Dr. Freitas is a contracted investigator at the Research Unit on Applied Molecular Biosciences (UCIBIO@REQUIMTE) in the Faculty of Pharmacy of the University of Porto, Portugal. She is currently the secretary of the Food- and Water-borne Infections Study Group from the European Society of Clinical Microbiology and Infectious Diseases. Her main research interests are in the molecular epidemiology, genomics, and evolution of antimicrobial-resistant Enterococcus.
Acknowledgment
This work was supported by the Applied Molecular Biosciences Unit—UCIBIO, which is financed by national funds from Fundação para a Ciência e Tecnologia (UIDP/04378/2020 and UIDB/04378/2020) and by the AgriFood XXI I&D&I project (NORTE-01-0145-FEDER-000041) cofinanced by European Regional Development Fund (ERDF) through the NORTE 2020 (Programa Operacional Regional do Norte 2014/2020). A.R.F. gratefully acknowledges the junior research position (CEECIND/02268/2017, Individual Call to Scientific Employment Stimulus 2017) granted by FCT/MCTES through national funds, and A.P.T. was supported by the Sara Borrell Research Grant (no. CD018/0123) funded by Instituto de Salud Carlos III and co-financed by the European Development Regional Fund (A Way to Achieve Europe program).
References
- Davies RH, Lawes JR, Wales AD. Raw diets for dogs and cats: a review, with particular reference to microbiological hazards. J Small Anim Pract. 2019;60:329–39. DOIPubMedGoogle Scholar
- van den Bunt G, Top J, Hordijk J, de Greeff SC, Mughini-Gras L, Corander J, et al. Intestinal carriage of ampicillin- and vancomycin-resistant Enterococcus faecium in humans, dogs and cats in the Netherlands. J Antimicrob Chemother. 2018;73:607–14. DOIPubMedGoogle Scholar
- Wu Y, Fan R, Wang Y, Lei L, Feßler AT, Wang Z, et al. Analysis of combined resistance to oxazolidinones and phenicols among bacteria from dogs fed with raw meat/vegetables and the respective food items. Sci Rep. 2019;9:15500. DOIPubMedGoogle Scholar
- Bender JK, Cattoir V, Hegstad K, Sadowy E, Coque TM, Westh H, et al. Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: Towards a common nomenclature. Drug Resist Updat. 2018;40:25–39. DOIPubMedGoogle Scholar
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. EUCAST version 10.0; 2020 [cited 2020 Dec 1]. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: twenty-eighth informational supplement M100. Annapolis Junction (MD): The Institute; 2018.
- Freitas AR, Tedim AP, Novais C, Lanza VF, Peixe L. Comparative genomics of global optrA-carrying Enterococcus faecalis uncovers a common chromosomal hotspot for optrA acquisition within a diversity of core and accessory genomes. Microb Genom. 2020;6:
e000350 . DOIPubMedGoogle Scholar - Novais C, Tedim AP, Lanza VF, Freitas AR, Silveira E, Escada R, et al. Co-diversification of Enterococcus faecium core genomes and PBP5: evidences of pbp5 horizontal transfer. Front Microbiol. 2016;7:1581. DOIPubMedGoogle Scholar
- Raven KE, Reuter S, Gouliouris T, Reynolds R, Russell JE, Brown NM, et al. Genome-based characterization of hospital-adapted Enterococcus faecalis lineages. Nat Microbiol. 2016;1:15033. DOIGoogle Scholar
- Zou J, Tang Z, Yan J, Liu H, Chen Y, Zhang D, et al. Dissemination of linezolid resistance through sex pheromone plasmid transfer in Enterococcus faecalis. Front Microbiol. 2020;11:1185. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: June 22, 2021
1These authors were co–principal investigators.
2These authors are active EFWISG members.
Table of Contents – Volume 27, Number 8—August 2021
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Luísa Peixe, UCIBIO, Departamento de Ciências Biológicas, Laboratório de Microbiologia, Faculdade de Farmácia, Universidade do Porto, Rua Jorge de Viterbo Ferreira, n. 228, 4050-313 Porto, Portugal
Top