Transmission of SARS-CoV-2 from Human to Domestic Ferret
Jožko Račnik

, Ana Kočevar, Brigita Slavec, Miša Korva, Katarina Resman Rus, Samo Zakotnik, Tomaž Mark Zorec, Mario Poljak, Milan Matko, Olga Zorman Rojs, and Tatjana Avšič Županc
Author affiliations: University of Ljubljana Faculty of Veterinary Medicine, Ljubljana, Slovenia (J. Račnik, B. Slavec, O. Zorman Rojs); Toplica Veterinary Hospital, Topolšica, Slovenia (A. Kočevar, M. Matko); University of Ljubljana Faculty of Medicine, Ljubljana (M. Korva, K. Resman Rus, S. Zakotnik, T.M. Zorec, M. Poljak, T. Avšič Županc)
Main Article
Figure 1
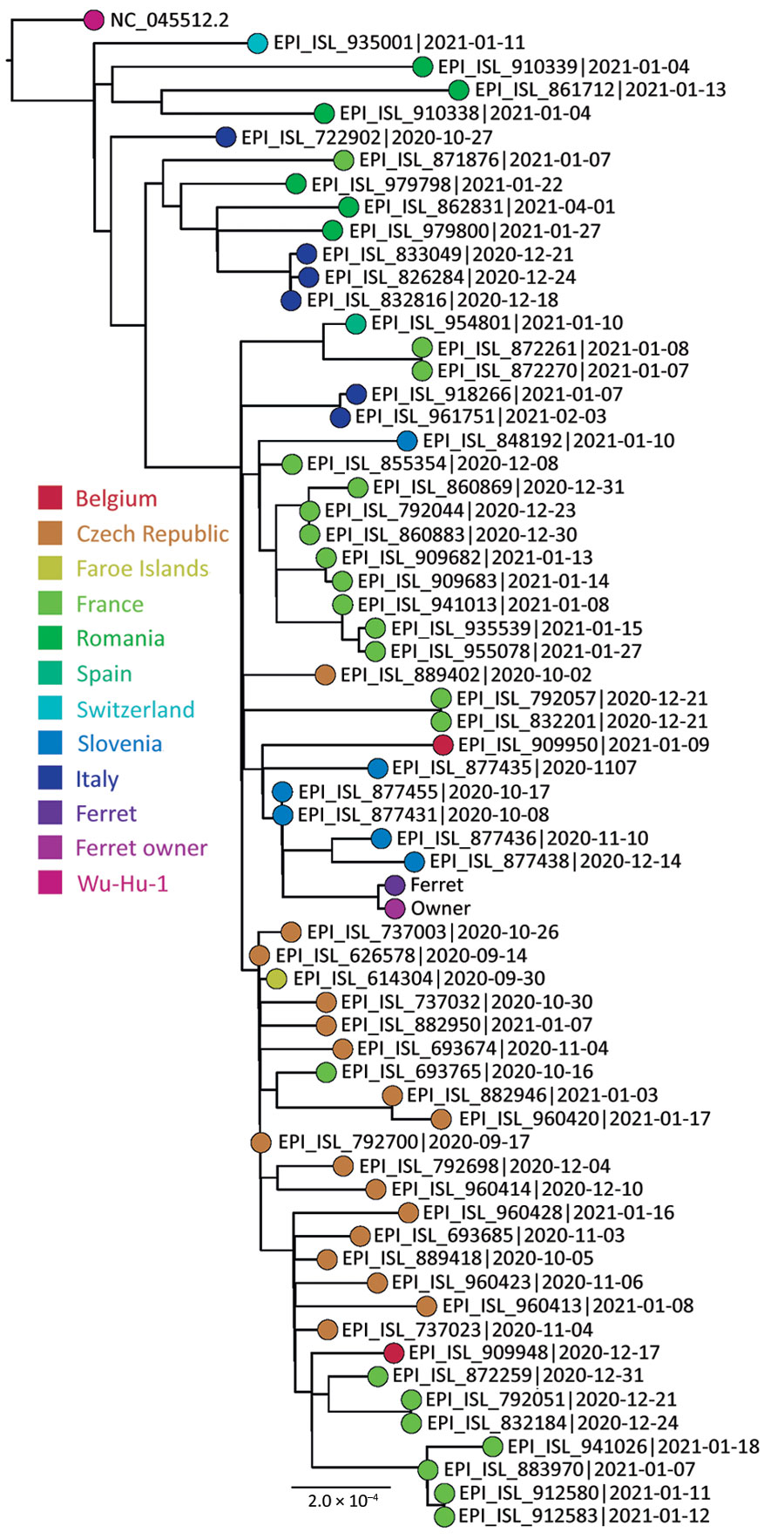
Figure 1. Phylogenetic sequence context consisted of high-quality complete severe acute respiratory syndrome coronavirus 2 genome sequences from a domestic ferret, Slovenia, corresponding to Pango lineage B.1.258. The context sequences were downloaded from GISAID (https://www.gisaid.org) and subsampled to 62 sequences and National Center for Biotechnology Information reference sequence NC_045512.2. The phylogenetic reconstruction using a general time-reversible plus F plus R4 substitution model was built in Figtree (Evomics, http://evomics.org) with 1,000 bootstrap replicates. The reference sequence was used as an outgroup to root the phylogenetic tree. GISAID accession numbers and isolation dates are provided. Scale bar indicates substitutions per site.
Main Article
Page created: July 13, 2021
Page updated: August 19, 2021
Page reviewed: August 19, 2021
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
