Volume 28, Number 1—January 2022
Research
High-Level Quinolone-Resistant Haemophilus haemolyticus in Pediatric Patient with No History of Quinolone Exposure
Figure
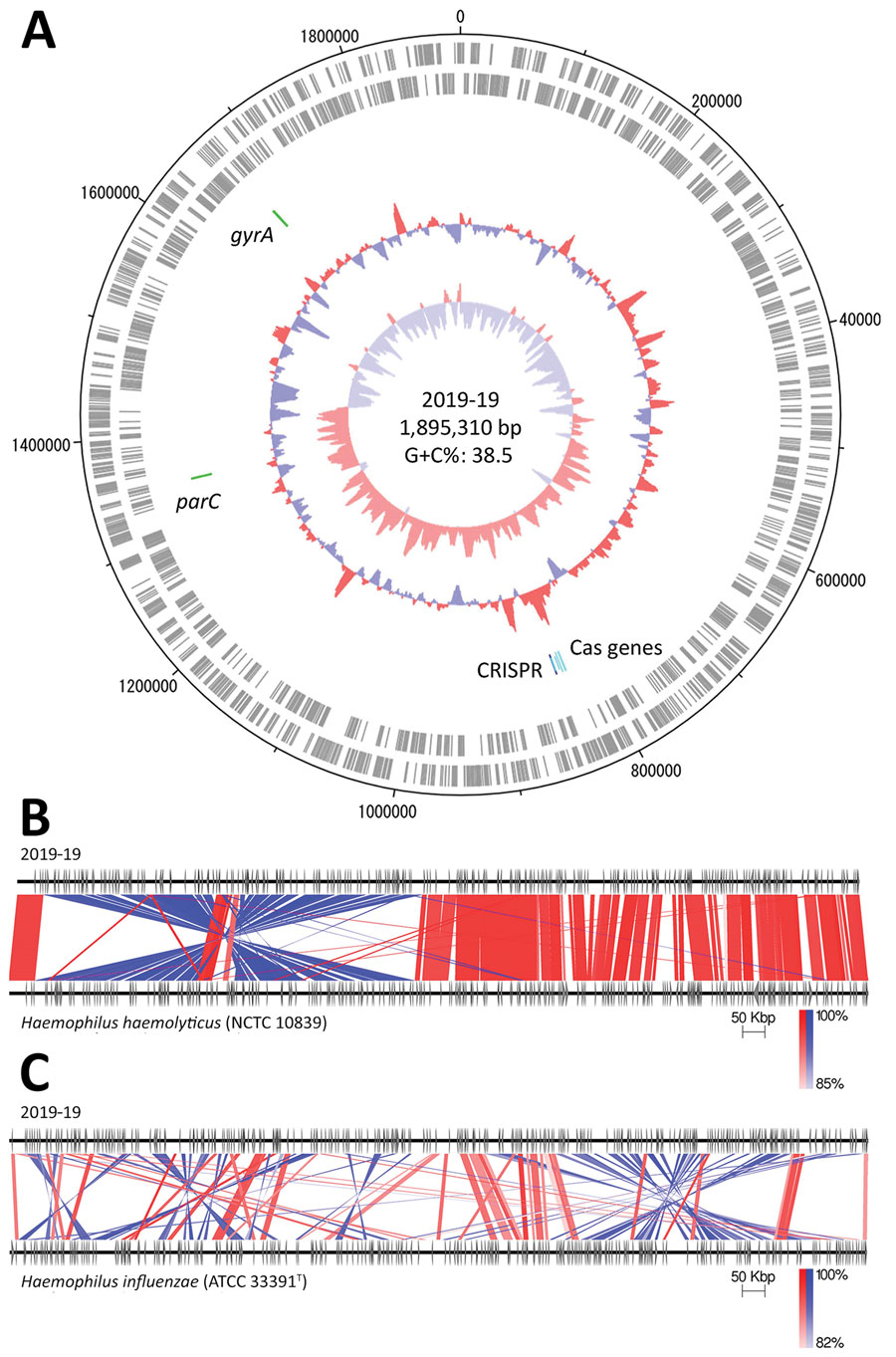
Figure. Genomic analysis of Haemophilus haemolyticus strain 2019-19 from a 9-year-old girl in Japan. A) Circular map of the whole-genome sequence. The outermost circle shows the number of nucleotides, the second circle shows coding sequences on the plus strand, and the third circle shows coding sequences on the minus strand. The innermost circle represents the G+C skew (%) and second innermost circle, G+C content (%); green zones show the locations of gyrA and parC, and blue and light blue zones show CRISPR-Cas–associated genes. Map drawn using Artemis DNA Plotter (Wellcome Sanger institute, Hinxton, UK). G+C, guanine + cytosine. B, C) Comparison between the whole genomes of 2019-19 and H. haemolyticus NCTC 10839 (B) and H. influenzae ATCC 33391T (C), created using Easyfig version 2.2.2 (19). Red indicates matches in the same direction; blue indicates inverted matches; white areas indicate nonmatches.
References
- Nørskov-Lauritsen N. Classification, identification, and clinical significance of Haemophilus and Aggregatibacter species with host specificity for humans. Clin Microbiol Rev. 2014;27:214–40. DOIPubMedGoogle Scholar
- Adachi Y, Ando M, Morozumi M, Ubukata K, Iwata S. Genotypic characterization of Haemophilus influenzae isolates from paediatric patients in Japan. J Med Microbiol. 2018;67:695–701. DOIPubMedGoogle Scholar
- Kilian M. Genus III. Haemophilus. Winslow, Broadhurst, Buchanan, Rogers and Smith 1917. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, editors. Bergey’s manual of systematic bacteriology, 2nd ed., vol. 2. The proteobacteriaceae. Part B. The gammaproteobacteria. New York: Springer; 2005. p. 883–904.
- Murphy TF, Brauer AL, Sethi S, Kilian M, Cai X, Lesse AJ. Haemophilus haemolyticus: a human respiratory tract commensal to be distinguished from Haemophilus influenzae. J Infect Dis. 2007;195:81–9. DOIPubMedGoogle Scholar
- Kirkham LA, Wiertsema SP, Mowe EN, Bowman JM, Riley TV, Richmond PC. Nasopharyngeal carriage of Haemophilus haemolyticus in otitis-prone and healthy children. J Clin Microbiol. 2010;48:2557–9. DOIPubMedGoogle Scholar
- McCrea KW, Xie J, LaCross N, Patel M, Mukundan D, Murphy TF, et al. Relationships of nontypeable Haemophilus influenzae strains to hemolytic and nonhemolytic Haemophilus haemolyticus strains. J Clin Microbiol. 2008;46:406–16. DOIPubMedGoogle Scholar
- Mukundan D, Ecevit Z, Patel M, Marrs CF, Gilsdorf JR. Pharyngeal colonization dynamics of Haemophilus influenzae and Haemophilus haemolyticus in healthy adult carriers. J Clin Microbiol. 2007;45:3207–17. DOIPubMedGoogle Scholar
- Hotomi M, Kono M, Togawa A, Arai J, Takei S, Ikeda Y, et al. Haemophilus influenzae and Haemophilus haemolyticus in tonsillar cultures of adults with acute pharyngotonsillitis. Auris Nasus Larynx. 2010;37:594–600. DOIPubMedGoogle Scholar
- Seyama S, Wajima T, Yanagisawa Y, Nakaminami H, Ushio M, Fujii T, et al. Rise in Haemophilus influenzae with reduced quinolone susceptibility and development of a simple screening method. Pediatr Infect Dis J. 2017;36:263–6. DOIPubMedGoogle Scholar
- Tanaka E, Hara N, Wajima T, Ochiai S, Seyama S, Shirai A, et al. Emergence of Haemophilus influenzae with low susceptibility to quinolones and persistence in tosufloxacin treatment. J Glob Antimicrob Resist. 2019;18:104–8. DOIPubMedGoogle Scholar
- Yokota S, Ohkoshi Y, Sato K, Fujii N. Emergence of fluoroquinolone-resistant Haemophilus influenzae strains among elderly patients but not among children. J Clin Microbiol. 2008;46:361–5. DOIPubMedGoogle Scholar
- Cherkaoui A, Gaïa N, Baud D, Leo S, Fischer A, Ruppe E, et al. Molecular characterization of fluoroquinolones, macrolides, and imipenem resistance in Haemophilus influenzae: analysis of the mutations in QRDRs and assessment of the extent of the AcrAB-TolC-mediated resistance. Eur J Clin Microbiol Infect Dis. 2018;37:2201–10. DOIPubMedGoogle Scholar
- Puig C, Tirado-Vélez JM, Calatayud L, Tubau F, Garmendia J, Ardanuy C, et al. Molecular characterization of fluoroquinolone resistance in nontypeable Haemophilus influenzae clinical isolates. Antimicrob Agents Chemother. 2015;59:461–6. DOIPubMedGoogle Scholar
- Tateda K, Ohno A, Ishii Y, Murakami H, Yamaguchi K; Levofloxacin surveillance group. Investigation of the susceptibility trends in Japan to fluoroquinolones and other antimicrobial agents in a nationwide collection of clinical isolates: A longitudinal analysis from 1994 to 2016. J Infect Chemother. 2019;25:594–604. DOIPubMedGoogle Scholar
- Yamada S, Seyama S, Wajima T, Yuzawa Y, Saito M, Tanaka E, et al. β-Lactamase-non-producing ampicillin-resistant Haemophilus influenzae is acquiring multidrug resistance. J Infect Public Health. 2020;13:497–501. DOIPubMedGoogle Scholar
- Ishiwada N, Fujimaki K, Matsumoto T, Kiyota H, Tateda K, Sato J, et al. Nationwide surveillance of bacterial pathogens isolated from children conducted by the surveillance committee of Japanese Society of Chemotherapy, the Japanese Association for Infectious Diseases, and the Japanese Society for Clinical Microbiology in 2017: General overview of pathogenic antimicrobial susceptibility. J Infect Chemother. 2021;27:139–50. DOIPubMedGoogle Scholar
- Marti S, Puig C, de la Campa AG, Tirado-Velez JM, Tubau F, Domenech A, et al. Identification of Haemophilus haemolyticus in clinical samples and characterization of their mechanisms of antimicrobial resistance. J Antimicrob Chemother. 2016;71:80–4. DOIPubMedGoogle Scholar
- Clinical Laboratory and Standards Institute. Performance standards for antimicrobial susceptibility testing (M100). 29th ed. Wayne (PA): The Institute; 2019.
- Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–10. DOIPubMedGoogle Scholar
- Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–31. DOIPubMedGoogle Scholar
- Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. DOIPubMedGoogle Scholar
- Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35(Web Server issue):W52–7.
- Christensen H, Kuhnert P, Olsen JE, Bisgaard M. Comparative phylogenies of the housekeeping genes atpD, infB and rpoB and the 16S rRNA gene within the Pasteurellaceae. Int J Syst Evol Microbiol. 2004;54:1601–9. DOIPubMedGoogle Scholar
- Bruin JP, Kostrzewa M, van der Ende A, Badoux P, Jansen R, Boers SA, et al. Identification of Haemophilus influenzae and Haemophilus haemolyticus by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Eur J Clin Microbiol Infect Dis. 2014;33:279–84. DOIPubMedGoogle Scholar
- Saffert RT, Cunningham SA, Ihde SM, Jobe KE, Mandrekar J, Patel R. Comparison of Bruker Biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometer to BD Phoenix automated microbiology system for identification of gram-negative bacilli. J Clin Microbiol. 2011;49:887–92. DOIPubMedGoogle Scholar
- Collins S, Vickers A, Ladhani SN, Flynn S, Platt S, Ramsay ME, et al. Clinical and molecular epidemiology of childhood invasive nontypeable Haemophilus influenzae disease in England and Wales. Pediatr Infect Dis J. 2016;35:e76–84. DOIPubMedGoogle Scholar
- Lulitanond A, Chanawong A, Pienthaweechai K, Sribenjalux P, Tavichakorntrakool R, Wilailuckana C, et al. Prevalence of β-lactamase-negative ampicillin-resistant haemophilus influenzae isolated from patients of a teaching hospital in Thailand. Jpn J Infect Dis. 2012;65:122–5.PubMedGoogle Scholar
- Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–31. DOIPubMedGoogle Scholar
- Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. DOIPubMedGoogle Scholar
- Harris TM, Price EP, Sarovich DS, Nørskov-Lauritsen N, Beissbarth J, Chang AB, et al. Comparative genomic analysis identifies X-factor (haemin)-independent Haemophilus haemolyticus: a formal re-classification of ‘Haemophilus intermedius’. Microb Genom. 2020;6:
e000303 . DOIPubMedGoogle Scholar - Georgiou M, Muñoz R, Román F, Cantón R, Gómez-Lus R, Campos J, et al. Ciprofloxacin-resistant Haemophilus influenzae strains possess mutations in analogous positions of GyrA and ParC. Antimicrob Agents Chemother. 1996;40:1741–4. DOIPubMedGoogle Scholar
- Chang CM, Tang HJ, Wang LR, Shih HI, Huang CC, Lee NY, et al. Increasing resistance to fluoroquinolones among Haemophilus species in Southern Taiwan. J Microbiol Immunol Infect. 2017;50:258–60. DOIPubMedGoogle Scholar
- Rodríguez-Martínez JM, López-Hernández I, Pascual A. Molecular characterization of high-level fluoroquinolone resistance in a clinical isolate of Haemophilus parainfluenzae. J Antimicrob Chemother. 2011;66:673–5. DOIPubMedGoogle Scholar
- Mikasa K, Aoki N, Aoki Y, Abe S, Iwata S, Ouchi K, et al. JAID/JSC guidelines for the treatment of respiratory infectious diseases: the Japanese Association for Infectious Diseases/Japanese Society of Chemotherapy—the JAID/JSC guide to clinical management of infectious disease/Guideline-preparing Committee Respiratory Infectious Disease WG. J Infect Chemother. 2016;22(Suppl):S1–65. DOIPubMedGoogle Scholar
- Wouters I, Desmet S, Van Heirstraeten L, Herzog SA, Beutels P, Verhaegen J, et al. NPcarriage Study Group. How nasopharyngeal pneumococcal carriage evolved during and after a PCV13-to-PCV10 vaccination programme switch in Belgium, 2016 to 2018. Euro Surveill. 2020;25:
1900303 . DOIGoogle Scholar - Takahata S, Ida T, Senju N, Sanbongi Y, Miyata A, Maebashi K, et al. Horizontal gene transfer of ftsI, encoding penicillin-binding protein 3, in Haemophilus influenzae. Antimicrob Agents Chemother. 2007;51:1589–95. DOIPubMedGoogle Scholar
- Witherden EA, Bajanca-Lavado MP, Tristram SG, Nunes A. Role of inter-species recombination of the ftsI gene in the dissemination of altered penicillin-binding-protein-3-mediated resistance in Haemophilus influenzae and Haemophilus haemolyticus. J Antimicrob Chemother. 2014;69:1501–9. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.