Volume 28, Number 1—January 2022
Research
Invasive Multidrug-Resistant emm93.0 Streptococcus pyogenes Strain Harboring a Novel Genomic Island, Israel, 2017–2019
Cite This Article
Citation for Media
Abstract
Invasive group A Streptococcus (iGAS) infections have increased in Israel since 2016 as successful lineages have emerged. We report the emergence and outbreak of a multidrug-resistant S. pyogenes emm93.0, sequence type 10, among iGAS infections in Israel since 2017. This type has been observed very rarely in other countries. During this period, emm93.0 was the cause of 116 infections in Israel and became the leading type during 2018. Most of the infections were from bacteremia (75%), and most patients were male (76%). We observed infections across Israel, mainly in adults. Of note, we observed multidrug resistance for clindamycin, tetracycline, and trimethoprim/sulfamethoxazole. Whole-genome sequencing confirmed clonality among geographically disseminated isolates. The local emm93.0 sequence type 10 clone contained a novel genomic island harboring the resistance genes lsa(E), lnu(B), and ant (6)-Ia aph(3′)-III. Further phenotypic and genomic studies are required to determine the prevalence of this resistance element in other iGAS types.
Streptococcus pyogenes is a major cause of community-acquired and nosocomial infections linked with illness and death worldwide (1,2). Group A Streptococcus (GAS) species cause a variety of infections, including pharyngitis, skin and soft tissue infections (SSTI), severe invasive infections, bacteremia, and toxic shock syndrome (3, 4). Acquisition of GAS is mainly attributed to person-to-person transmission by respiratory droplets or skin contact; in addition, inoculated food can spread the infection, resulting in outbreaks (5,6).
Surveillance programs and prevention guidelines focus on systemic GAS infection, which is defined as a statutory notifiable disease in many countries (5). Active laboratory-based surveillance based on molecular characterization of invasive isolates is essential for outbreak detection and public health response. However, laboratory surveillance data indicate that most invasive cases in industrialized countries occur sporadically and are not part of outbreak clusters (5). In Israel, iGAS is a notifiable disease; all invasive isolates are analyzed at the national reference laboratory at the Ministry of Health (Jerusalem, Israel).
The emm typing scheme is a primary tool for surveillance, outbreak detection, and for the study of the population structure on the basis of the sequencing of the emm gene (7). Specific emm types and M proteins (M1, M3) have been linked with invasive infections. Several studies show that emm types are significantly more diverse in developing countries than in developed countries (5).
Susceptibility testing is essential for successful outbreak control. Although β-lactams are the preferred antimicrobial drug treatment for GAS infections, macrolides, lincosamides, and streptogramins are useful for treating patients with β-lactam allergy, and for overcoming treatment failure in patients treated with penicillin. Recommendations for treating severe invasive cases include adding clindamycin or linezolid, which suppress toxin production (8,9). Several reports highlight the emergence of successful clones associated with acquired antimicrobial resistance (10–12). Clindamycin resistance is rarely reported and is associated with specific lineages (13,14).
We describe the epidemiology of invasive GAS (iGAS) in Israel during 2014–2019. We report the emergence and ongoing outbreak of a multidrug-resistant (MDR) S. pyogenes emm93.0 that caused 116 iGAS cases during 2016–2019 in Israel. The unique epidemiologic dynamics of the outbreak clone, as well as in-depth whole-genome sequence analysis, were the focus of our investigation.
Strain Typing and Susceptibility Testing
In Israel, all S. pyogenes strains isolated from normally sterile sites are referred to the national Streptococcus reference center as part of routine surveillance. All isolates are cultured on blood agar base plates (HyLabs, https://www.hylabs.co.il) or in Todd-Hewitt broth at 37°C. We conducted a survey of 66 noninvasive GAS strains from throat and ear samples received from 3 medical centers to detect carriage of emm93.0. We subjected all S. pyogenes isolates to Lancefield grouping (15) and to emm typing in accordance with the guidelines stated by the US Centers for Disease Control and Prevention (CDC; https://www2.cdc.gov/vaccines/biotech/strepblast.asp).
We determined the antibiotic susceptibility of emm93.0 strains by broth microdilution using Sensititer (TREK Diagnostic Systems, https://www.trekds.com) with STP6F antimicrobial susceptibility test (AST) plates containing 20 antimicrobial drugs, according to the manufacturer’s instructions. We used Sensititre Vizion (Thermo Fisher, https://www.thermofisher.com) for manual reading of growth. We interpreted MIC according to Clinical and Laboratory Standards Institute (CLSI) 2019 guidelines except for trimethoprim/sulfamethoxazole, for which CLSI breakpoints were unavailable; therefore, we used the epidemiologic cutoff value, 0.5 μg/mL, from the European Committee on Antimicrobial Susceptibility Testing (https://www.eucast.org/clinical_breakpoints).
Whole-Genome Sequencing Analysis
Of 116 emm93.0 strains, we subjected 26 (22.4%), representing different isolation dates and geographic locations, to whole-genome sequencing (Appendix 1 Table 2). We performed DNA extraction using the QIAsymphony SP system and the QIAsymphony DNA mini kit (QIAGEN, https://www.qiagen.com) according to the manufacturer’s recommendations. We lysed culture pellets in a 180 µL enzymatic lysis buffer (20 mM Tris-Cl pH 8.0 (VWR Amresco, https://us.vwr.com), 2 mM sodium EDTA (VWR Amresco), 1.2% Triton X-100 (Sigma-Aldrich, https://www.sigmaaldrich.com) and lysozyme 20 mg/mL (Sigma-Aldrich). We incubated cell suspension for 30 min at 37°C, then treated with proteinase K and buffer AL (QIAGEN). We subjected silica beads in lysing matrix B bulk (MP Biomedicals, https://www.mpbio.com), in a volume equivalent to 150 μL, to the TissueLyser II (QIAGEN) for 2 cycles of 30 Hz for 30 s. We transferred supernatants to the QIAsymphony SP system.
We sequenced Nextera XT DNA Libraries (Illumina, https://www.illumina.com) on MiSeq (Illumina), using a read length of 250 bp paired-end at >100× coverage. We analyzed reads by the BioNumerics version 7.6.3 (Applied Maths, https://www.applied-maths.com). We generated de novo assemblies using SPAdes version 3.7.1 (https://github.com/ablab/spades/tree/spades_3.7.1). To determine clonality of the outbreak strains, we mapped reads (Appendix 1 Table 2) against a reference strain (IST003) using the bowtie algorithm. We used the BioNumerics Gene Extraction tool (Applied Maths) to search the assemblies for sequences of antimicrobial resistance genes and virulence factors, in accordance with published gene sequences (Appendix 1 Table 3) (16). To identify the genetic basis for antimicrobial resistance, we analyzed the genomes using the Pathosystems Resource Integration Center (PATRIC) version 3.6.6 (17). We obtained visualizations for comparison of IST001, IST003, and GAS2887HUB (Sequence Read Archive [SRA] accession no. ERR2880947) putative prophage insertion site using the Easyfig program (18). We used Fisher exact test for statistical analysis and comparison of categorical variables between groups.
Epidemiology of iGAS
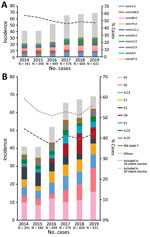
During 2014–2019, a total of 2,947 iGAS isolates were analyzed at the Streptococcus reference laboratory (Figure 1, panel A). The most common source of iGAS was blood (51.3%), followed by wounds (21.7%). The incidence of iGAS was 40–70 cases/100,000 population/year. We observed an increasing trend in incidence during 2016–2019 (Figure 1, panel A). A total of 180 emm types were identified; the 10 leading types were emm1.0 (n = 307, 10.4%), emm106.0 (n = 215, 7.3%), emm89.0 (n = 189, 6.4%), emm75.0 (n = 136, 4.6%), emm112.2 (n = 123, 4.2%), emm93.0 (n = 116, 3.9%), emm12.0 (n = 104, 3.5%), emm22.0 (n = 102, 3.5%), emm4.0 (n = 90, 3.0%), and emm87.0 (n = 84, 2.8%). The predominant type in Israel was the globally reported emm1.0 that caused 9.7%–11.4% of iGAS cases per year; it ranked first or second in each year (19). The top 10 emm types of each year accounted for 52%–60% of total iGAS cases (Appendix 1 Table 1). Among the 10 leading emm types, some remained stable throughout the studied period, with few exceptions (Figure 1; Appendix 1 Table 1). The 180 emm types identified during 2014–2019 were clustered into 38 emm acceptable clusters (20). The top 10 emm clusters accounted for >86% of total cases; they were E4 (N = 590, 20%), E6 (N = 343, 11.6%), A-C3 (N = 333, 11.3%), E3 (N = 287, 9.7%), E2 (E = 257, 8.7%), D4 (E = 247, 8.4%), E1 (N = 152, 5.2%), A-C4 (N = 117, 4%), A-C5 (N = 114, 3.9%), and M6 clade Y (N = 104, 3.5%) (Figure 1, panel B). The largest emm cluster, E4, peaked in 2019. It represented <20% of iGAS cases, and included mainly types emm89.0, emm112.2, and emm22.0. Cluster D4 was rarely detected during 2014–2016 (28 cases), but peaked in 2017–2019 (219 cases). The predominant types among cluster D4 were emm93.0 (47%), emm53.3 (24.7%), and emm33.0 (20.2%).
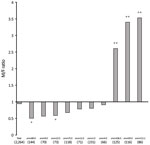
The potential coverage of multivalent vaccines was low in Israel compared with other industrialized countries (21). The yearly predicted coverage of the 26-valent S. pyogenes vaccine was 36.7%–44.6% of the invasive strains, and 40.7% for the entire period. The yearly predicted coverage of the 30-valent S. pyogenes vaccine was 51%–59.2% of invasive strains, and 53.7% for the entire period (Figure 1, panel B).
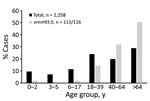
During 2016–2019, a total of 2,263 iGAS cases were distributed almost evenly between sexes (48.2% male, 51.8% female). However, differences in sex of patients were noticeable in several emm types (Figure 2). Most patients (77%) with emm93.0 were male; emm106.0 and emm112.2 had significantly higher incidence (p<0.05) in male patients (Figure 2). In contrast, emm89.0 and emm9.0 were significantly more prevalent (p<0.05) in female patients (Figure 2). Most iGAS cases (72.2%) were reported in adults >17 years of age, and the most affected age group was >64 years of age (28.7%) (Figure 3).
emm93.0 Emergence and Outbreak
Our surveillance data highlight the emergence of emm93.0 during 2016, followed by an ongoing outbreak across Israel of this rarely reported type. After a single case in April 2016, emm93.0 emerged during 2017 and caused 4.3% of cases, 9.8% of cases in 2018, and 4.9% of cases in 2019. A total of 116 emm93.0 invasive GAS cases were diagnosed during April 2016–December 2019; cases peaked in October in 2017–2019 (Figure 4). The isolates were recovered mainly from blood (76% of total cases) and wound specimens. Thirteen (52%) of the isolates were from blood and 11 (44%) from wounds in 2017. In 2018, bacteremia cases yielded 51 (86%) isolates (Figure 4). Of note, in a survey of 66 throat and ear samples from the community, emm93.0 was not detected among noninvasive cases (Appendix 1 Table 4). Most emm93.0 cases were among adult patients from age groups 39–64 years (31%) and >64 years of age (51%), versus 20% of total iGAS cases from patients 39–64 years of age and 29% from those >64 years of age (Figure 3).
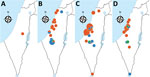
We noted the geographic distribution of 106/116 (91.4%) emm93.0 cases (Figure 5). The earliest reported case of emm93.0 in Israel was in a patient from Judea and Samaria district. During 2017–2019, cases were reported from 17 medical centers and disseminated across Israel. The outbreak clone (82% of total emm93.0 cases) was distributed mainly in 3/7 districts of Israel: South, Tel-Aviv, and Center (Figure 5).
Antimicrobial Resistance
The outbreak clones were notably resistant to clindamycin (MIC >1 μg/mL), tetracycline (MIC >8 μg/mL), and trimethoprim/sulfamethoxazole (MIC >4 μg/mL). All outbreak isolates were susceptible to azithromycin, cefepime, cefotaxime, ceftriaxone, chloramphenicol, daptomycin, ertapenem, erythromycin, levofloxacin, linezolid, meropenem, moxifloxacin, penicillin, and vancomycin. The earliest-reported emm93.0 isolate identified in Israel, IST001, was resistant to tetracycline (MIC >8 μg/mL) and trimethoprim/sulfamethoxazole (MIC >4 μg/mL) but susceptible to clindamycin (MIC ≤0.12 μg/mL) and all other antimicrobial drugs tested.
Whole-Genome Sequencing Analysis
We investigated a sample of 26 isolates (22.4%) by whole-genome sequencing to confirm clonality and characterize the emerging type (Appendix 1 Table 3). We used the first known isolate from the southern district of Israel (IST003) as a reference sequence. Sequence mapping confirmed clonality of the outbreak strains isolated during 2017–2019, identifying <11 single-nucleotide polymorphism (SNPs). SNPs were accumulated during the outbreak years: <4 SNPs in 2017, <6 SNPs in 2018, and <11 SNPs in 2019 compared with the reference strain. We determined that IST001, the first emm93.0 isolate isolated in 2016, was not part of the outbreak from its high number of SNPs (>350 SNPs). SNPs were distributed throughout the genome; we observed 5 areas with dense SNPs (Appendix 1 Table 5; Appendix 2 Figure 1), which rules out the option of a single recombination event. We performed BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis to map and identify the genes located in these areas. Among those genes were genes coding for proteins with various functions such as metabolism and biosynthesis (LacA, LacB, GatA, GatB, GatC, FabK, CodY) and transport (PTS lactose transporters, ABC transporters, ECF transporters); proteins involved in DNA replication, recombination, transcription and translation (MutL, RecG, XRE family, MarR family, ribosomal proteins S9, L13); toxins (holin-like, type II toxin-antitoxin system); and several proteins with unknown functions (Appendix 1 Table 5).
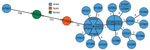
We extracted sequences of 7 housekeeping genes (gki, gtr, murI, mutS, recP, xpt, ypiL) from the assemblies and submitted them to PubMLST (https://pubmlst.org) to determine the sequence type (ST) of the strains. All strains were ST10 and had the allelic profile 2-2-9-13-2-14-2. ST10 was reported in only 12 strains in pubMLST from tropical and subtropical regions, associated with 7 rare emm types, 93, 70, 80, 98, 108, 121, and 142. To further analyze the strains, we conducted whole-genome multilocus sequence typing (wgMLST) analysis using the BioNumerics S. pyogenes scheme (Applied Maths). Among the outbreak strains, we detected <9 allelic differences (Figure 6). We compared the Israel strains to globally available sequences of emm93.0 and detected 20 allelic differences from GAS2887HUB (SRA accession no. ERR2880947) a strain reported in Spain (22), and 153 allelic differences from an isolate from Kenya, K40810 (SRA accession no. ERR227074) (23).
Antimicrobial-Resistance Genes and Genes Contributing To Virulence
We used PATRIC version 3.6.6 Specialty Genes Search tool to identify antimicrobial resistance genes in the genome of the resistant S. pyogenes emm93.0 isolate. We compared the search results of a resistant strain, IST003, to the sensitive strain, IST001; the genes found only in the resistant strain were ant(6)-I, aph(3′)-III lnu(B), and lsa(E). In addition, tet(M) was found in both strains. No amino acid replacement was found in PBP2x (24). To identify gene arrangement and their genomic environment, we searched gene sequences in de novo assembled genomes using BioNumerics platform tools. Using BLAST, we defined a genomic island 56,821-bp long, at IST003 positions 1427343–1484164. This region was identical to a genomic island harboring lsa(E), lnu(B), and a defective prophage in S. pyogenes emm93.0 strain, previously described in Spain (22). In the United States, the lsa/lnu type determinants are extremely rare in invasive GAS, but are not uncommon among invasive group B Streptococcus (21,25). The genomic island presumably integrated into the rlmD gene (Figure 7).
Among the SAg genes, we found speB, speG, speM and smeZ in all outbreak strains; we detected slo as well. The first emm93.0 clone, isolated in Israel (IST001) has a different pattern of genes: speB, speC, speG, speH, speI, speM, smeZ, slo, and tetM (Appendix 2 Figure 2; Appendix 1 Table 3).
We observed an increase in incidence of iGAS in Israel from 2014 to 2019 (40–70 cases/100,000 population). The iGAS population in Israel displayed high genetic diversity. Global data reflect limited diversity in industrialized countries, and higher diversity in developing countries. The consistently high iGAS diversity in Israel, an industrialized country, is noteworthy and concerning. The potential coverage of multivalent emm-type–specific vaccines is expected to be limited in Israel compared with vaccine coverage in Europe and the United States. In Israel the strain types included in the protein-based 26-valent vaccines cover 40.7% of cases and in the 30-valent vaccines, 53.7% of cases, compared with 79% coverage by the 26-valent vaccines and 91% of cases by the 30-valent vaccines in the United States (26).
The molecular surveillance of all iGAS cases reflects trends and fluctuations. The leading strain type in Israel during 2014–2019 was emm1.0, similar to global reports. Of note, emm93.0 has never been detected in the ongoing molecular surveillance of iGAS initiated in 2003. This type was not detected in a sample of noninvasive GAS, and the source and reservoir of this clone are currently unknown. Because cluster D was found to be associated with impetigo and skin infections (27,28), it could be related to skin carriage. In the United States, outbreak clusters were associated with increasingly more invasive GAS (emm types 49, 82, and 92) (21).
The emm93.0 type has been reported very rarely in other countries. In Spain, a single case of iGAS in 2009 was caused by an MDR emm93.0 isolate carrying a defective prophage (22). Most characterized S. pyogenes exotoxins, deoxyribonucleases, or DNases are carried by prophages, which contribute to its virulence, immune evasion, and bacterial DNA degradation (29). We compared the Israel outbreak’s WGS profile with the case in Spain and report a resemblance, with 20 allelic differences identified by wgMLST. These differences were scattered in different genomic regions and not likely due to a single recombination event. The reported resistance phenotype and genotype of the Spain isolate were identical to those of the outbreak cluster in Israel. The prevalence of this type in Spain is currently unknown. A study of GAS in New Caledonia reported 18 isolates of emm93 among 318 cases (5.6%) from 2012 (30); a single invasive case was reported, but subtype and antimicrobial susceptibility were not reported.
The emergence and outbreak of an MDR clone among iGAS cases is concerning in 2 ways. First, many patients with SSTI and a history of β-lactam allergy receive empiric treatment with clindamycin, but results of antimicrobial susceptibility testing are unknown for many of these patients because cultures from SSTIs are not commonly positive. Second, patients with invasive streptococcal infections and hemodynamic instability are commonly treated with clindamycin to inhibit toxin production. If iGAS clones with clindamycin resistance become prevalent, a different empiric approach might be considered, such as the use of linezolid.
The emerging emm93.0 type is not covered by the multivalent vaccines under development. The high genetic similarity of the Israel outbreak clone to a single case from Spain from 2009 may indicate an epidemiologic link and global transmission. Systematic molecular surveillance of iGAS is essential for detection of local and global emerging clones and for evidence-based vaccine development and distribution.
Dr. Ron is head of the National Streptococcus Reference Laboratory at the Ministry of Health, Israel. Her interests include genomic epidemiology and infectious diseases.
Acknowledgment
The laboratory work of this study was performed at the Public Health Laboratory, Jerusalem, Israel, and supported by the Israel Ministry of Health. We thank Amichai Ron for his assistance with generating Figures 5 and 7.
References
- Ralph AP, Carapetis JR. Group a streptococcal diseases and their global burden. Curr Top Microbiol Immunol. 2013;368:1–27.PubMedGoogle Scholar
- Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–94. DOIPubMedGoogle Scholar
- Barnett TC, Bowen AC, Carapetis JR. The fall and rise of Group A Streptococcus diseases. Epidemiol Infect. 2018;147:
e4 . DOIPubMedGoogle Scholar - Henningham A, Barnett TC, Maamary PG, Walker MJ. Pathogenesis of group A streptococcal infections. Discov Med. 2012;13:329–42.PubMedGoogle Scholar
- Efstratiou A, Lamagni T. Epidemiology of Streptococcus pyogenes. In: Ferretti JJ, Stevens DL, Fischetti VA, editors. Streptococcus pyogenes: basic biology to clinical manifestations. Oklahoma City (OK): University of Oklahoma Health Sciences Center; 2016.
- Kemble SK, Westbrook A, Lynfield R, Bogard A, Koktavy N, Gall K, et al. Foodborne outbreak of group a streptococcus pharyngitis associated with a high school dance team banquet—Minnesota, 2012. Clin Infect Dis. 2013;57:648–54. DOIPubMedGoogle Scholar
- Smeesters PR, Botteaux A. The emm-cluster typing system. Methods Mol Biol. 2020;2136:25–31. DOIPubMedGoogle Scholar
- Diep BA, Equils O, Huang DB, Gladue R. Linezolid effects on bacterial toxin production and host immune response: review of the evidence. Curr Ther Res Clin Exp. 2012;73:86–102. DOIPubMedGoogle Scholar
- Zimbelman J, Palmer A, Todd J. Improved outcome of clindamycin compared with beta-lactam antibiotic treatment for invasive Streptococcus pyogenes infection. Pediatr Infect Dis J. 1999;18:1096–100. DOIPubMedGoogle Scholar
- Oppegaard O, Skrede S, Mylvaganam H, Kittang BR. Emerging threat of antimicrobial resistance in β-hemolytic streptococci. Front Microbiol. 2020;11:797. DOIPubMedGoogle Scholar
- Tsai WC, Shen CF, Lin YL, Shen FC, Tsai PJ, Wang SY, et al. Emergence of macrolide-resistant Streptococcus pyogenes emm12 in southern Taiwan from 2000 to 2019. J Microbiol Immunol Infect. 2020;•••:
S1684-1182(20)30217-6 ; Epub ahead of print. DOIPubMedGoogle Scholar - Brouwer S, Barnett TC, Ly D, Kasper KJ, De Oliveira DMP, Rivera-Hernandez T, et al. Prophage exotoxins enhance colonization fitness in epidemic scarlet fever-causing Streptococcus pyogenes. Nat Commun. 2020;11:5018. DOIPubMedGoogle Scholar
- Pesola AK, Sihvonen R, Lindholm L, Pätäri-Sampo A. Clindamycin resistant emm33 Streptococcus pyogenes emerged among invasive infections in Helsinki metropolitan area, Finland, 2012 to 2013. Euro Surveill. 2015;20:21117. DOIPubMedGoogle Scholar
- Richter SS, Heilmann KP, Beekmann SE, Miller NJ, Miller AL, Rice CL, et al. Macrolide-resistant Streptococcus pyogenes in the United States, 2002-2003. Clin Infect Dis. 2005;41:599–608. DOIPubMedGoogle Scholar
- Lancefield RC. The antigenic complex of Streptococcus haemolyticus: I. Demonstration of a type-specific substance in extracts of Streptococcus haemolyticus. J Exp Med. 1928;47:91–103. DOIPubMedGoogle Scholar
- Turner CE, Holden MTG, Blane B, Horner C, Peacock SJ, Sriskandan S. The emergence of successful Streptococcus pyogenes lineages through convergent pathways of capsule loss and recombination directing high toxin expression. MBio. 2019;10:e02521–19. DOIPubMedGoogle Scholar
- Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42(D1):D581–91. DOIPubMedGoogle Scholar
- Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–10. DOIPubMedGoogle Scholar
- Nir-Paz R, Korenman Z, Ron M, Michael-Gayego A, Cohen-Poradosu R, Valinsky L, et al. Streptococcus pyogenes emm and T types within a decade, 1996-2005: implications for epidemiology and future vaccines. Epidemiol Infect. 2010;138:53–60. DOIPubMedGoogle Scholar
- Sanderson-Smith M, De Oliveira DM, Guglielmini J, McMillan DJ, Vu T, Holien JK, et al.; M Protein Study Group. A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J Infect Dis. 2014;210:1325–38. DOIPubMedGoogle Scholar
- Li Y, Rivers J, Mathis S, Li Z, Velusamy S, Nanduri SA, et al. Genomic surveillance of Streptococcus pyogenes strains causing invasive disease, United States, 2016–2017. Front Microbiol. 2020;11:1547. DOIPubMedGoogle Scholar
- Berbel D, Càmara J, García E, Tubau F, Guérin F, Giard JC, et al. A novel genomic island harbouring lsa(E) and lnu(B) genes and a defective prophage in a Streptococcus pyogenes isolate resistant to lincosamide, streptogramin A and pleuromutilin antibiotics. Int J Antimicrob Agents. 2019;54:647–51. DOIPubMedGoogle Scholar
- Seale AC, Davies MR, Anampiu K, Morpeth SC, Nyongesa S, Mwarumba S, et al. Invasive group A Streptococcus infection among children, rural Kenya. Emerg Infect Dis. 2016;22:224–32. DOIPubMedGoogle Scholar
- Musser JM, Beres SB, Zhu L, Olsen RJ, Vuopio J, Hyyryläinen HL, et al. Reduced in vitro susceptibility of Streptococcus pyogenes to β-lactam antibiotics associated with mutations in the pbp2x gene is geographically widespread. J Clin Microbiol. 2020;58:e01993–19. DOIPubMedGoogle Scholar
- McGee L, Chochua S, Li Z, Mathis S, Rivers J, Metcalf B, et al. Multistate, population-based distributions of candidate vaccine targets, clonal complexes, and resistance features of invasive group B streptococci within the United States, 2015–2017. Clin Infect Dis. 2021;72:1004–13. DOIPubMedGoogle Scholar
- Gherardi G, Vitali LA, Creti R. Prevalent emm types among invasive GAS in Europe and North America since year 2000. Front Public Health. 2018;6:59. DOIPubMedGoogle Scholar
- Bessen DE, Carapetis JR, Beall B, Katz R, Hibble M, Currie BJ, et al. Contrasting molecular epidemiology of group A streptococci causing tropical and nontropical infections of the skin and throat. J Infect Dis. 2000;182:1109–16. DOIPubMedGoogle Scholar
- Bessen DE. Tissue tropisms in group A Streptococcus: what virulence factors distinguish pharyngitis from impetigo strains? Curr Opin Infect Dis. 2016;29:295–303. DOIPubMedGoogle Scholar
- Jespersen MG, Lacey JA, Tong SYC, Davies MR. Global genomic epidemiology of Streptococcus pyogenes. Infect Genet Evol. 2020;86:
104609 . DOIPubMedGoogle Scholar - Baroux N, D’Ortenzio E, Amédéo N, Baker C, Ali Alsuwayyid B, Dupont-Rouzeyrol M, et al. The emm-cluster typing system for Group A Streptococcus identifies epidemiologic similarities across the Pacific region. Clin Infect Dis. 2014;59:e84–92. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: December 08, 2021
Table of Contents – Volume 28, Number 1—January 2022
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Assaf Rokney, Ministry of Health, Central Laboratories, Eliav 9, Jerusalem 91342, Israel
Top