Volume 28, Number 11—November 2022
Dispatch
Sequence-Based Identification of Metronidazole-Resistant Clostridioides difficile Isolates
Figure
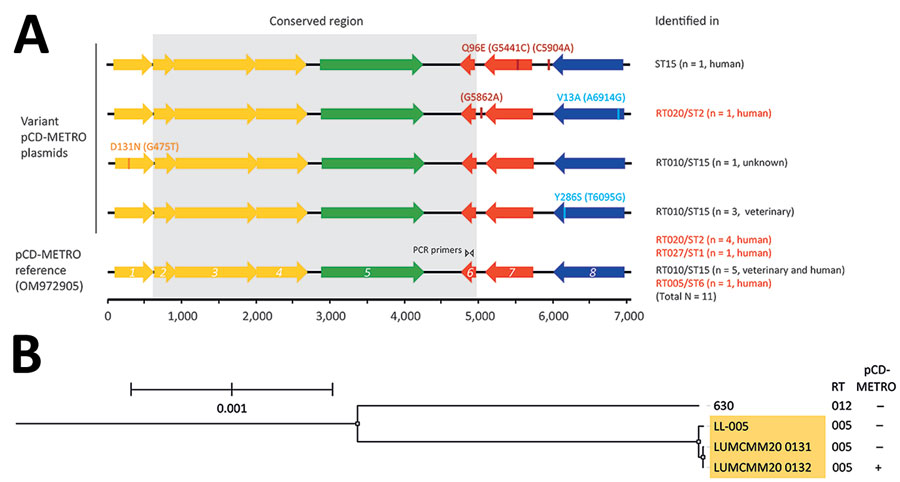
Figure. Comparison of pCD-METRO open reading frames and phylogenetic analysis in study of sequence-based identification of metronidazole-resistant Clostridioides difficile isolates. A) Linear maps compare the open reading frames (ORF)1–8 of the pCD-METRO reference sequence (identical to the RT005 plasmid) with variant pCD-METRO sequences, including the ST15 isolate from the United States (top). No ribotyping information was available for the ST15 isolate, but it should be noted that RT010 isolates belong to the same sequence type. Amino acid substitutions and nucleotide substitutions (in parentheses) are indicated above the ORFs. Colors indicate the location of putative mobilization genes (yellow), a replication gene (green), an integrase gene (blue), and genes encoding other functions (red) in the ORFs (3). The invariant regions are indicated by gray shading, and the binding location of the oBH1/2 primer set is shown in ORF6. The primer set is used for national sentinel surveillance and diagnostics of C. difficile infections in the Netherlands. Toxigenic RT/STs are indicated in red font and were all derived from symptomatic patients with C. difficile infections. Where available, the source (human/veterinary) is indicated. Isolate 1143 from Brazil was not included in this figure because no sequence information was available. B) Phylogenetic tree generated using IQ-TREE (10) and Roary (11) to show the relatedness between 2 RT005 patient isolates (LUMCMM20 0131 and LUMCMM20 0132) compared with the 2 reference strains LL-005 (RT005) and 630 (RT012). The tree is rooted on strain 630, and RT005 isolates are highlighted in yellow. Only the LUMCMM20 0132 isolate was positive for pCD-METRO. Scale bar indicates nucleotide substitutions per site. RT, ribotype; ST, sequence type.
References
- Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. Clostridium difficile infection. Nat Rev Dis Primers. 2016;2:16020. DOIPubMedGoogle Scholar
- Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, et al.; Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399:629–55. DOIPubMedGoogle Scholar
- Boekhoud IM, Hornung BVH, Sevilla E, Harmanus C, Bos-Sanders IMJG, Terveer EM, et al. Plasmid-mediated metronidazole resistance in Clostridioides difficile. Nat Commun. 2020;11:598. DOIPubMedGoogle Scholar
- Boekhoud IM, Sidorov I, Nooij S, Harmanus C, Bos-Sanders IMJG, Viprey V, et al.; COMBACTE-CDI Consortium. Haem is crucial for medium-dependent metronidazole resistance in clinical isolates of Clostridioides difficile. J Antimicrob Chemother. 2021;76:1731–40. DOIPubMedGoogle Scholar
- Moura I, Spigaglia P, Barbanti F, Mastrantonio P. Analysis of metronidazole susceptibility in different Clostridium difficile PCR ribotypes. J Antimicrob Chemother. 2013;68:362–5. DOIPubMedGoogle Scholar
- Albuquerque C, Pagnossin D, Landsgaard K, Simpson J, Brown D, Irvine J, et al. The duration of antibiotic treatment is associated with carriage of toxigenic and non-toxigenic strains of Clostridioides difficile in dogs. PLoS One. 2021;16:
e0245949 . DOIPubMedGoogle Scholar - Freeman J, Vernon J, Pilling S, Morris K, Nicholson S, Shearman S, et al.; Pan-European Longitudinal Surveillance of Antibiotic Resistance among Prevalent Clostridium difficile Ribotypes Study Group. The ClosER study: results from a three-year pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes, 2011-2014. Clin Microbiol Infect. 2018;24:724–31. DOIPubMedGoogle Scholar
- Arkin AP, Cottingham RW, Henry CS, Harris NL, Stevens RL, Maslov S, et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat Biotechnol. 2018;36:566–9. DOIPubMedGoogle Scholar
- Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. DOIPubMedGoogle Scholar
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. DOIPubMedGoogle Scholar
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3. DOIPubMedGoogle Scholar
- Blackwell GA, Hunt M, Malone KM, Lima L, Horesh G, Alako BTF, et al. Exploring bacterial diversity via a curated and searchable snapshot of archived DNA sequences. PLoS Biol. 2021;19:
e3001421 . DOIPubMedGoogle Scholar - Worley J, Delaney ML, Cummins CK, DuBois A, Klompas M, Bry L. Genomic determination of relative risks for Clostridioides difficile infection from asymptomatic carriage in intensive care unit patients. Clin Infect Dis. 2021;73:e1727–36. DOIPubMedGoogle Scholar
- Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1):D517–25.PubMedGoogle Scholar
- Carattoli A, Hasman H. PlasmidFinder and In Silico pMLST: identification and typing of plasmid replicons in whole-genome sequencing (WGS). Methods Mol Biol. 2020;2075:285–94. DOIPubMedGoogle Scholar