Volume 28, Number 4—April 2022
Research Letter
Legionella pneumophila Subspecies fraseri Infection after Allogeneic Hematopoietic Stem Cell Transplant, China
Cite This Article
Citation for Media
Abstract
We describe an immunosuppressed patient with bacteremia and pneumonia caused by Legionella pneumophila subspecies fraseri in China. We confirmed this diagnosis by using nanopore sequencing of positive blood cultures and subsequent recovery from buffered-charcoal yeast extract culture. Nanopore sequencing is an effective tool for early diagnosis of atypical infections.
Legionella pneumophila is an opportunistic atypical pathogen of community-acquired or hospital-acquired pneumonia (1–3). Underestimates of its prevalence are likely because the Legionella urinary antigen testing and buffered-charcoal yeast extract (BCYE) culture routinely used for diagnosis are sometimes available only in a few tertiary hospitals that specialize in respiratory diseases. Therefore, early diagnosis and prompt therapy for L. pneumophila infection are crucial. We report a case of L. pneumophila subspecies fraseri bloodstream infection in a patient in China after allogeneic hematopoietic stem cell transplant (aHSCT). We confirmed this diagnosis by using nanopore sequencing of positive blood cultures and by recovering L. pneumofila using BCYE medium. Chest radiography and computed tomography (CT) suggested acute L. pneumophila pneumonia. The study was approved by the Institutional Review Board of the Peking University People’s Hospital (approval no. 2019PHB134–01).
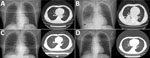
A 55-year-old man with acute T/myeloid mixed-cell leukemia was hospitalized because of nasal bleeding on day 62 after aHSCT. After treatment for thrombocytopenia, he experienced mild diarrhea on day 101. Three days later (day 104), his temperature was 38.8°C (Appendix Figure 1), but he had no accompanying symptoms. One aerobic blood culture vial, 1 anaerobic blood culture vial, and 1 Myco/F blood culture vial (BD, https://www.bd.com) per puncture point (2 puncture points) were collected for culture. Chest radiography and CT displayed multiple solid nodules in both lungs; the largest was in the right lower lobe, which suggested infection (Figure). The patient then experienced a 3-day continuous high fever with a maximum temperature of 39.5°C, a small amount of white sputum, and a blood oxygen saturation of 90% on day 107; the same number of blood culture vials were collected. CT showed a large, solid, fuzzy shadow with a unclear boundary, uneven internal density, and low-density plaques in the lower lobe of the right lung (Figure). The patient was administered linezolid (0.6 g every 12 h) and imipenem (0.5 g every 8 h) to control the infection. The patient’s temperature decreased to a normal level, but the C-reactive protein values did not decrease to reference range (Appendix Figure 2).
After ≈10 days (256 h) of incubation, we observed that the blood cultures in the Myco/F vial collected on day 104 after aHSCT, which are commonly used to culture Mycobacterium spp. and fungus, contained the only growing gram-negative bacilli. We then transferred the positive blood cultures to blood nutrition plates for further culture in 5% carbon dioxide, anaerobic, and microaerobic environments, but this culture failed. Simultaneously, we directly used a serum separator-gel tube and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (4) to identify positive blood cultures but were unsuccessful. Subsequently, we extracted DNA from the positive blood cultures for nanopore sequencing (5,6); L. pneumophila subsp. fraseri was detected after 1 hour (Appendix; BioProject Short Read Archive accession no. PRJNA744850). The coverage length and depth were 37.67% and 2.04 with 1,337 raw reads. After timely communication of the sequencing results to the clinical department, the patient was administered azithromycin (0.5 g 1×/d for 1 d) , followed by moxifloxacin (0.4 g 1×/d for 9 d) to ease symptoms of discomfort caused by azithromycin. The infection was finally controlled, and diarrhea symptoms improved 3 days after appropriate moxifloxacin therapy was initiated.
Afterward, we analyzed serum collected on May 24 (day 114) and May 27 (day 117) and detected L. pneumophila IgM by using an ELISA kit (Euroimmun, https://www.euroimmun.com). Legionella urinary antigen testing was not performed. Meanwhile, 100 μL of the positive blood cultures was inoculated on the in-house BCYE agar supplied with Legionella BCYE growth supplement medium (OXOID; Thermo Fisher Scientific, https://www.thermofisher.com). Two days later, we successfully isolated L. pneumophila and displayed wet blue-purple luster colonies (Appendix Figure 3); L. pneumophila was finally identified using MALDI-TOF mass spectrometry. During the course of infection, no more sputum specimens were available because the patient had no obvious cough or expectoration.
The fatality rate of Legionnaires’ disease is between 5% and 30% (7). Risk factors for L. pneumophila infection include age >50 years, solid tumors or hematologic malignancies, solid organ transplant, and immunosuppression (8,9). Hence, shortening the turnaround time to identify microorganisms is crucial for timely diagnosis and appropriate therapy, which influence death rates. In this case, the diagnosis of L. pneumophila pneumonia was not possible on the basis of the atypical radiologic evidence, high fever (>38°C), elevated C-reactive protein, and diarrhea, although such manifestations are the most common symptoms of L. pneumophila infections (10). Furthermore, administration of corticosteroids and immunosuppressive drugs likely obscured the respiratory symptoms. Rapid nanopore sequencing with a short turnaround time has the potential to effectively expedite the detection of L. pneumophila infection, guaranteeing the appropriate antibiotic therapy, especially for immunosuppressed patients with atypical symptoms.
Dr. Wang is a researcher in the Department of Clinical Laboratory, Peking University People’s Hospital, Beijing, China, focusing on the detection and study of resistance mechanisms of antimicrobial agents. Mr. Yifan Guo is a PhD candidate under the supervision of Dr. Wang in the Institute of Medical Technology, Peking University Health Science Center and the Department of Clinical Laboratory, Peking University People’s Hospital, focusing on metagenomic sequencing and bioinformatics analysis.
Acknowledgments
We thank Zhenzhong Li and Hongbin Chen for bioinformatics analysis, Ying Shang and Zhengwu Yang for serologic testing for L. pneumophila IgM, Jingzhi Wang for clinical communication and consultation, and Feifei Zhang for preparing of BYCE culture medium.
This work was supported by the National Key Research and Development Program of China (2018YFE0102100 and 2018YFC1200102).
References
- Gonçalves IG, Simões LC, Simões M. Legionella pneumophila. Trends Microbiol. 2021;29:860–1. DOIPubMedGoogle Scholar
- Miyashita N, Higa F, Aoki Y, Kikuchi T, Seki M, Tateda K, et al. Distribution of Legionella species and serogroups in patients with culture-confirmed Legionella pneumonia. J Infect Chemother. 2020;26:411–7. DOIPubMedGoogle Scholar
- Qin T, Ren H, Chen D, Zhou H, Jiang L, Wu D, et al. National surveillance of Legionnaires’ disease, China, 2014-2016. Emerg Infect Dis. 2019;25:1218–9. DOIPubMedGoogle Scholar
- Yonetani S, Ohnishi H, Ohkusu K, Matsumoto T, Watanabe T. Direct identification of microorganisms from positive blood cultures by MALDI-TOF MS using an in-house saponin method. Int J Infect Dis. 2016;52:37–42. DOIPubMedGoogle Scholar
- Charalampous T, Kay GL, Richardson H, Aydin A, Baldan R, Jeanes C, et al. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol. 2019;37:783–92. DOIPubMedGoogle Scholar
- Moon J, Jang Y, Kim N, Park WB, Park KI, Lee ST, et al. Diagnosis of Haemophilus influenzae pneumonia by Nanopore 16S Amplicon sequencing of sputum. Emerg Infect Dis. 2018;24:1944–6. DOIPubMedGoogle Scholar
- Edelstein PH, Roy CR. Legionella. In: Bennett JE, Dolin R, Blaser MJ, editors. Mandell, Douglas, and Bennett’s principles and practice of infectious diseases, 8th ed. Philadelphia (PA); Elsevier; 2014. p. 2633–44.
- Pouderoux C, Ginevra C, Descours G, Ranc AG, Beraud L, Boisset S, et al. Slowly or nonresolving Legionnaires’ disease: case series and literature review. Clin Infect Dis. 2020;70:1933–40. DOIPubMedGoogle Scholar
- Cunha BA, Burillo A, Bouza E. Legionnaires’ disease. Lancet. 2016;387:376–85. DOIPubMedGoogle Scholar
- Phin N, Parry-Ford F, Harrison T, Stagg HR, Zhang N, Kumar K, et al. Epidemiology and clinical management of Legionnaires’ disease. Lancet Infect Dis. 2014;14:1011–21. DOIPubMedGoogle Scholar
Figure
Cite This ArticleOriginal Publication Date: March 11, 2022
1These authors contributed equally to this article.
Table of Contents – Volume 28, Number 4—April 2022
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Hui Wang, Department of Clinical Laboratory, Peking University People’s Hospital, Xicheng District, Beijing, 100044, China
Top