Volume 28, Number 6—June 2022
Synopsis
Detection of SARS-CoV-2 B.1.351 (Beta) Variant through Wastewater Surveillance before Case Detection in a Community, Oregon, USA
Figure 1
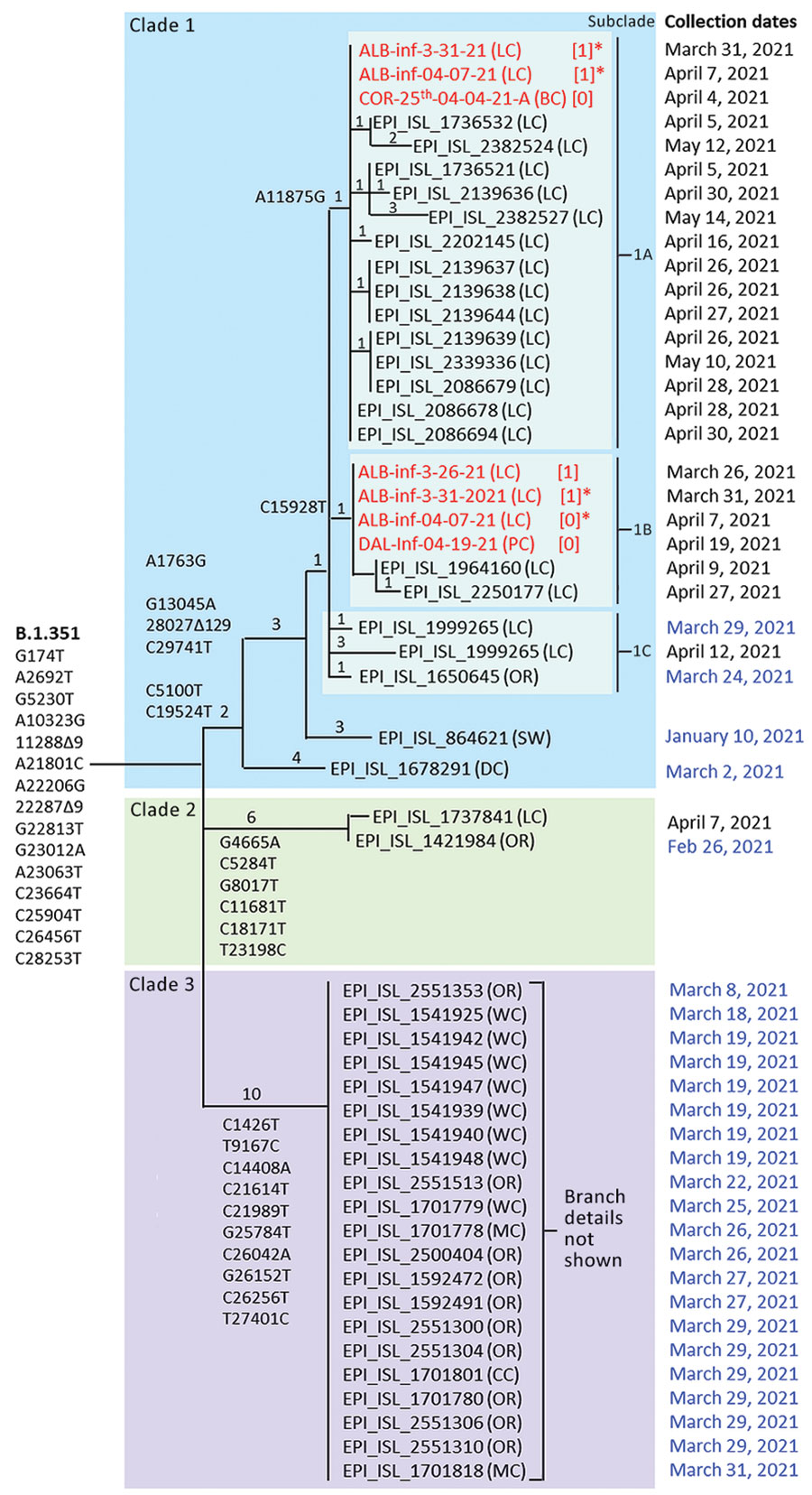
Figure 1. Maximum-parsimony tree demonstrating phylogenetic relationships among SARS-CoV-2 variant B.1.351 clinical specimens and wastewater samples in Linn County, Oregon, USA, and surrounding jurisdictions, March–May 2021. GISAID accession numbers (https://www.gisaid.org) are shown for 19 of 20 B.1.351 specimens identified in Linn County through May 15, 2021, and for 24 additional B.1.351 specimens identified in Oregon through March 31, 2021 (dates in blue). Also included are 2 sequences from outside Oregon (Switzerland and Washington, DC, USA) most closely related to clade 1. Wastewater samples are in red. Exact parsimony trees are shown for clade 1 and 2 sequences, whereas clade 3 sequences are simply listed. Mutations defining B.1.351 and each of the 3 clades, plus subclades 1a and 1b, are shown. Private mutations defining the subbranches of clades 1 and 2 are listed in Appendix Table 9. Numbers on tree branches indicate the numbers of mutations associated with each branch. Numbers in brackets indicate clade 1 consensus mutations not detected, probably because of poor read coverage. Asterisks indicate samples that appear in both subclades 1a and 1b and are inferred to be a mixture of at >2 B.1.351 subtypes. Wastewater sequences ALB-Inf-04-21-21-A and COR-26th-04-04-21-A are not shown because several tracts of those sequences were too uncertain to enable accurate placement on the tree. OR, Oregon; BC, Benton County; CC, Clackamas County; LC, Linn County; MC, Multnomah County; WC, Washington County; DC, Washington, DC; SW, Switzerland.